4. Additional information and examples#
Tutorial at the 2024 paleoCAMP | June 18–July 1, 2024
Jiang Zhu
jiangzhu@ucar.edu
Climate & Global Dynamics Laboratory
NSF National Center for Atmospheric Research
More information and examples to demonstrate model data access and analysis using the NCAR JupyterHub
Further Info 1: Guidance on using climate data
Further Info 2: Access ESM output
Example 1: Access CESM output on NCAR’s Campaign Storage and perform model-data comparison
Example 2: Access ERA5 Reanalysis on NSF NCAR’s Research Data Archive
Example 3: Plot AMOC from TraCE and compare with McManus et al. 2004
Example 4. Compute and plot precipitation δ18O
Time to go through: 10 minutes
4.1. Further Info 1: Guidance on using climate data#
NCAR Climate Data Guide: Key strength, Key limitations, Expert User Guidance, etc.
Paleoclimate (You could contribute!)
…
4.2. Further Info 2: Access ESM output#
Earth System Grid Federation (ESGF), e.g., the LLNL Node
NCAR Climate Data Gateway, e.g., the iTRACE
NCAR Research Data Archive: e.g., EAR5 Reanalysis (1.77 PB!)
Other portals, such as the DeepMIP Model Database
NCAR JupyterHub is your
one-stop shopfor CESM data and analysis!Multiple PB of CESM Paleoclimate simulation data
Other CESM simulation data
Preinstalled Python environment
Note: variable names may be different depending on the portals (IPCC Standard; CESM Standard)
Load Python packages
import os
import glob
from datetime import timedelta
import xarray as xr
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
from cartopy.util import add_cyclic_point
# xesmf is used for regridding ocean output
import xesmf
import warnings
warnings.filterwarnings("ignore")
4.3. Example 1: Access CESM paleoclimate output and perform model-data comparison on NCAR’s Campaign Storage#
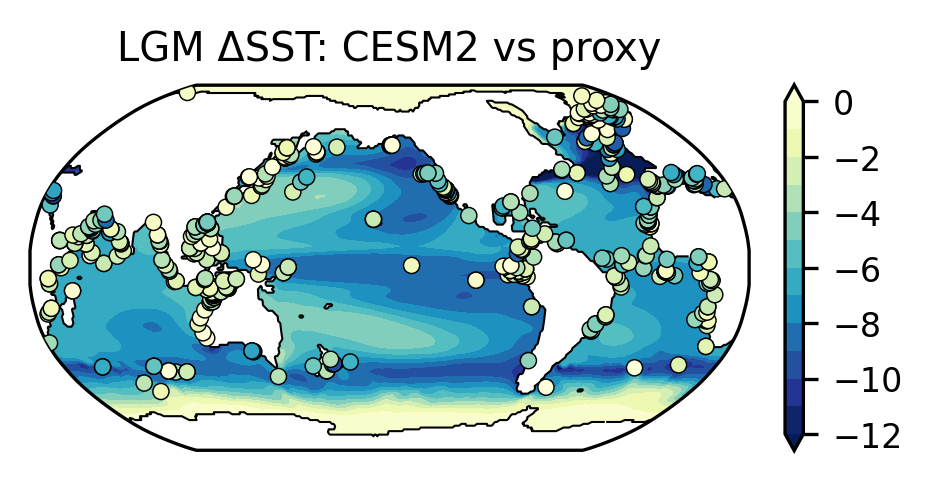
We try to reproduce Figure 2a of Jiang’s paper to use the LGM ΔSST to assess CESM2’s climate sensitivity.
Whole set of model output is shared at:
/glade/campaign/cesm/development/palwgYou can find additional simulation data here:
/glade/campaign/cgd/ppc/jiangzhuWe are in the process of organizing available data on the Paleoclimate Working Group webiste
As for now, prior knowledge of the experiments and file structure are needed. We hope to develop a Simulation Catalog for this!
!ls /glade/campaign/cesm/development/palwg/
cesm Idealized old.setups
CMIP_DECK inputdata paleoweather
ctsm_glacier_gridfiles_LGM LastGlacialMaximum pliocene
datasets LastInterglacial Pliocene
EXTRA LastMillennium raw_boundary_data
holocene lig-H11 Tabor_paleo
Holocene LIGtransient temp
Holocene-9ka mvr
!ls -l /glade/campaign/cesm/development/palwg/LastGlacialMaximum/CESM2
!ls /glade/campaign/cesm/development/palwg/LastGlacialMaximum/CESM2/b.e21.B1850CLM50SP.f09_g17.PI.01/ocn/proc/tseries/month_1/*.TEMP.*
!ls /glade/campaign/cesm/development/palwg/LastGlacialMaximum/CESM2/b.e21.B1850CLM50SP.f09_g17.21ka.01/ocn/proc/tseries/month_1/*.TEMP.*
total 3
drwxr-sr-x+ 7 jiangzhu cesm 4096 Feb 24 2023 b.e21.B1850CLM50SP.f09_g17.21ka.01
drwxr-sr-x+ 8 jiangzhu cesm 4096 Feb 24 2023 b.e21.B1850CLM50SP.f09_g17.PI.01
-rw-r-----+ 1 jiangzhu cesm 413 Aug 14 2023 please_cite_zhu_etal_2021
/glade/campaign/cesm/development/palwg/LastGlacialMaximum/CESM2/b.e21.B1850CLM50SP.f09_g17.PI.01/ocn/proc/tseries/month_1/b.e21.B1850CLM50SP.f09_g17.PI.01.pop.h.TEMP.000101-010012.nc
/glade/campaign/cesm/development/palwg/LastGlacialMaximum/CESM2/b.e21.B1850CLM50SP.f09_g17.PI.01/ocn/proc/tseries/month_1/b.e21.B1850CLM50SP.f09_g17.PI.01.pop.h.TEMP.010101-020012.nc
/glade/campaign/cesm/development/palwg/LastGlacialMaximum/CESM2/b.e21.B1850CLM50SP.f09_g17.PI.01/ocn/proc/tseries/month_1/b.e21.B1850CLM50SP.f09_g17.PI.01.pop.h.TEMP.020101-030012.nc
/glade/campaign/cesm/development/palwg/LastGlacialMaximum/CESM2/b.e21.B1850CLM50SP.f09_g17.21ka.01/ocn/proc/tseries/month_1/b.e21.B1850CLM50SP.f09_g17.21ka.01.pop.h.TEMP.000101-010012.nc
/glade/campaign/cesm/development/palwg/LastGlacialMaximum/CESM2/b.e21.B1850CLM50SP.f09_g17.21ka.01/ocn/proc/tseries/month_1/b.e21.B1850CLM50SP.f09_g17.21ka.01.pop.h.TEMP.010101-020012.nc
/glade/campaign/cesm/development/palwg/LastGlacialMaximum/CESM2/b.e21.B1850CLM50SP.f09_g17.21ka.01/ocn/proc/tseries/month_1/b.e21.B1850CLM50SP.f09_g17.21ka.01.pop.h.TEMP.020101-030012.nc
/glade/campaign/cesm/development/palwg/LastGlacialMaximum/CESM2/b.e21.B1850CLM50SP.f09_g17.21ka.01/ocn/proc/tseries/month_1/b.e21.B1850CLM50SP.f09_g17.21ka.01.pop.h.TEMP.030101-040012.nc
/glade/campaign/cesm/development/palwg/LastGlacialMaximum/CESM2/b.e21.B1850CLM50SP.f09_g17.21ka.01/ocn/proc/tseries/month_1/b.e21.B1850CLM50SP.f09_g17.21ka.01.pop.h.TEMP.040101-050012.nc
campaign_dir = '/glade/campaign/cesm/development/palwg/LastGlacialMaximum/CESM2'
comp = 'ocn/proc/tseries/month_1'
4.3.1. Read preindustrial SST#
case = 'b.e21.B1850CLM50SP.f09_g17.PI.01'
fname = 'b.e21.B1850CLM50SP.f09_g17.PI.01.pop.h.TEMP.020101-030012.nc'
file = os.path.join(campaign_dir, case, comp, fname)
# Open the file and select the last 10 years of data
ds_pre = xr.open_dataset(file).isel(time=slice(-120, None))
sst_pre = ds_pre.TEMP.isel(z_t=0).mean('time')
sst_pre
<xarray.DataArray 'TEMP' (nlat: 384, nlon: 320)>
array([[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
[-1.9042568, -1.9034731, -1.9024575, ..., nan, nan,
nan],
...,
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan],
[ nan, nan, nan, ..., nan, nan,
nan]], dtype=float32)
Coordinates:
z_t float32 500.0
ULONG (nlat, nlon) float64 ...
ULAT (nlat, nlon) float64 ...
TLONG (nlat, nlon) float64 ...
TLAT (nlat, nlon) float64 ...
Dimensions without coordinates: nlat, nlon4.3.2. Read LGM SST#
case = 'b.e21.B1850CLM50SP.f09_g17.21ka.01'
fname = 'b.e21.B1850CLM50SP.f09_g17.21ka.01.pop.h.TEMP.040101-050012.nc'
file = os.path.join(campaign_dir, case, comp, fname)
# Open the file and select the last 10 years of data
ds_lgm = xr.open_dataset(file).isel(time=slice(-120, None))
sst_lgm = ds_lgm.TEMP.isel(z_t=0).mean('time')
sst_lgm
<xarray.DataArray 'TEMP' (nlat: 384, nlon: 320)>
array([[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
...,
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan],
[nan, nan, nan, ..., nan, nan, nan]], dtype=float32)
Coordinates:
z_t float32 500.0
ULONG (nlat, nlon) float64 ...
ULAT (nlat, nlon) float64 ...
TLONG (nlat, nlon) float64 ...
TLAT (nlat, nlon) float64 ...
Dimensions without coordinates: nlat, nlon4.3.3. Regrid into the 1° × 1° grid using xesmf#
%%time
ds_pre['lat'] = ds_pre.TLAT
ds_pre['lon'] = ds_pre.TLONG
regridder = xesmf.Regridder(
ds_in=ds_pre,
ds_out=xesmf.util.grid_global(1, 1, cf=True, lon1=360),
method='bilinear',
periodic=True)
dsst_1x1 = regridder(sst_lgm - sst_pre)
dsst_1x1
CPU times: user 6.62 s, sys: 244 ms, total: 6.86 s
Wall time: 7.2 s
<xarray.DataArray (lat: 180, lon: 360)>
array([[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
[ nan, nan, nan, ..., nan,
nan, nan],
...,
[-0.17081444, -0.17075008, -0.17069343, ..., -0.17104828,
-0.17096627, -0.1708865 ],
[-0.178788 , -0.17878205, -0.17877994, ..., -0.17882909,
-0.17881154, -0.17879783],
[-0.18547155, -0.18547687, -0.18548317, ..., -0.18546143,
-0.18546383, -0.1854672 ]], dtype=float32)
Coordinates:
z_t float32 500.0
* lon (lon) float64 0.5 1.5 2.5 3.5 ... 357.5 358.5 359.5
latitude_longitude float64 nan
* lat (lat) float64 -89.5 -88.5 -87.5 -86.5 ... 87.5 88.5 89.5
Attributes:
regrid_method: bilinear4.3.4. Read proxy ΔSST in csv from Jess’s github#
url = 'https://raw.githubusercontent.com/jesstierney/lgmDA/master/proxyData/Tierney2020_ProxyDataPaired.csv'
proxy_dsst = pd.read_csv(url)
proxy_dsst.head()
| Latitude | Longitude | Lower2s | Median | Upper2s | ProxyType | Species | |
|---|---|---|---|---|---|---|---|
| 0 | -55.0 | 73.3 | -2.927296 | -1.379339 | 0.302709 | delo | pachy |
| 1 | -53.0 | -58.0 | 1.426191 | 2.773898 | 4.253553 | uk | NaN |
| 2 | -51.1 | 67.7 | -4.810437 | -3.192364 | -0.570439 | delo | pachy |
| 3 | -48.1 | 146.9 | -4.784523 | -3.370838 | -1.886123 | uk | NaN |
| 4 | -46.1 | 90.1 | -4.949764 | -3.470513 | -1.919271 | delo | bulloides |
4.3.5. Plot the LGM ΔSST in the model and proxy records#
cmap = plt.get_cmap('YlGnBu').reversed()
norm = mpl.colors.Normalize(-12, 0)
fig, ax = plt.subplots(nrows=1, ncols=1,
figsize=(3, 1.5),
subplot_kw={'projection': ccrs.Robinson(central_longitude=210)},
constrained_layout=True)
# Plot model results using contourf
dsst_1x1_new, lon_new = add_cyclic_point(dsst_1x1, dsst_1x1.lon)
p0 = ax.contourf(lon_new, dsst_1x1.lat, dsst_1x1_new,
levels=np.linspace(-12, 0, 13),
cmap=cmap, norm=norm, extend='both',
transform=ccrs.PlateCarree())
plt.colorbar(p0, ax=ax)
# Create a land-sea mask and plot the LGM coastal line
lmask = xr.where(dsst_1x1.isnull(), 0, 1)
ax.contour(lmask.lon, lmask.lat, lmask,
levels=[0.5],
linewidths=0.5,
colors='black',
transform=ccrs.PlateCarree())
# Plot proxy SST using markers
ax.scatter(proxy_dsst['Longitude'],
proxy_dsst['Latitude'],
c=proxy_dsst['Median'],
marker='o',
s=15,
cmap=cmap,
edgecolors='black',
lw=0.35,
norm=norm,
zorder=3,
transform=ccrs.PlateCarree())
ax.set_title("LGM ΔSST: CESM2 vs proxy")
Text(0.5, 1.0, 'LGM ΔSST: CESM2 vs proxy')
4.4. Example 2: Access ERA5 Reanalysis data on NSF NCAR’s Research Data Archive#
Browse the RDA website and figure out the file structure
Search
era5Click the
Monthly Meanproduct (ds633.1)Click the tab
DATA ACCESSTotal precipitation is in
ERA5 monthly mean atmospheric surface forecast (accumulated) [netCDF4]Click
GLADE File Listingto see all the files stored on the NCAR Campaign Storage
data_dir = '/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/'
files_tp = glob.glob(data_dir + '*/*_tp.*.nc')
print(*files_tp, sep='\n')
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2003/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2003010100_2003120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2018/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2018010100_2018120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2004/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2004010100_2004120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2019/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2019010100_2019120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2005/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2005010100_2005120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2006/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2006010100_2006120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1979/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1979010100_1979120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2014/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2014010100_2014120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2000/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2000010100_2000120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1987/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1987010100_1987120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2015/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2015010100_2015120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2001/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2001010100_2001120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1988/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1988010100_1988120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2016/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2016010100_2016120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2002/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2002010100_2002120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1989/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1989010100_1989120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2017/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2017010100_2017120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2010/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2010010100_2010120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1997/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1997010100_1997120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1983/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1983010100_1983120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2011/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2011010100_2011120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1998/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1998010100_1998120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1984/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1984010100_1984120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2012/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2012010100_2012120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1999/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1999010100_1999120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1985/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1985010100_1985120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2013/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2013010100_2013120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2020/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2020010100_2020120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1986/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1986010100_1986120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1993/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1993010100_1993120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2021/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2021010100_2021120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1994/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1994010100_1994120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2022/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2022010100_2022120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1980/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1980010100_1980120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1995/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1995010100_1995120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1981/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1981010100_1981120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1996/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1996010100_1996120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1982/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1982010100_1982120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2007/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2007010100_2007120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1990/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1990010100_1990120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2008/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2008010100_2008120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1991/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1991010100_1991120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/2009/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.2009010100_2009120100.nc
/glade/campaign/collections/rda/data/ds633.1/e5.moda.fc.sfc.accumu/1992/e5.moda.fc.sfc.accumu.128_228_tp.ll025sc.1992010100_1992120100.nc
ds = xr.open_mfdataset(files_tp)
ds = ds.reindex(latitude=sorted(ds.latitude.values))
ds
<xarray.Dataset>
Dimensions: (latitude: 721, longitude: 1440, time: 528)
Coordinates:
* latitude (latitude) float64 -90.0 -89.75 -89.5 -89.25 ... 89.5 89.75 90.0
* longitude (longitude) float64 0.0 0.25 0.5 0.75 ... 359.0 359.2 359.5 359.8
* time (time) datetime64[ns] 1979-01-01 1979-02-01 ... 2022-12-01
Data variables:
TP (time, latitude, longitude) float32 dask.array<chunksize=(3, 332, 776), meta=np.ndarray>
utc_date (time) int32 dask.array<chunksize=(12,), meta=np.ndarray>
Attributes:
DATA_SOURCE: ECMWF: https://cds.climate.copernicus.eu, Copernicu...
NETCDF_CONVERSION: CISL RDA: Conversion from ECMWF GRIB 1 data to netC...
NETCDF_VERSION: 4.6.1
CONVERSION_PLATFORM: Linux casper02 3.10.0-693.21.1.el7.x86_64 #1 SMP We...
CONVERSION_DATE: Mon Nov 11 08:45:33 MST 2019
Conventions: CF-1.6
NETCDF_COMPRESSION: NCO: Precision-preserving compression to netCDF4/HD...
history: Mon Nov 11 08:45:34 2019: ncks -4 --ppc default=7 e...
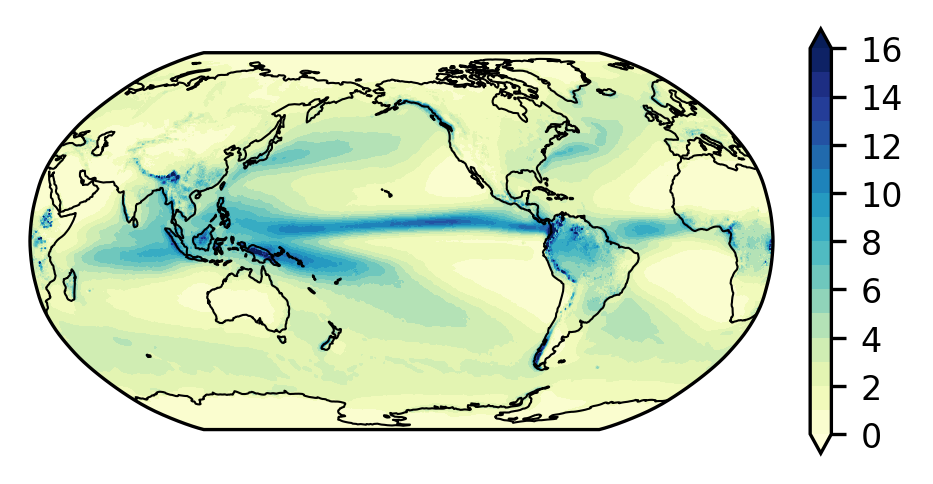
NCO: netCDF Operators version 4.7.9 (Homepage = http://n...4.4.1. Change the units from m per day to mm per day and make a plot#
See here for clarification of units.
tp = ds.TP.mean('time') * 1000.0
tp = tp.compute()
tp
<xarray.DataArray 'TP' (latitude: 721, longitude: 1440)>
array([[0.18871191, 0.18871191, 0.18871191, ..., 0.18871191, 0.18871191,
0.18871191],
[0.1763593 , 0.17630874, 0.17631234, ..., 0.17640808, 0.17640086,
0.17640446],
[0.17613353, 0.17612088, 0.17608838, ..., 0.17611367, 0.17612632,
0.17613533],
...,
[0.6987416 , 0.69886446, 0.6989421 , ..., 0.6986495 , 0.69864047,
0.69868386],
[0.7080526 , 0.7081086 , 0.70814294, ..., 0.70803094, 0.7080472 ,
0.70808154],
[0.7012306 , 0.7012306 , 0.7012306 , ..., 0.7012306 , 0.7012306 ,
0.7012306 ]], dtype=float32)
Coordinates:
* latitude (latitude) float64 -90.0 -89.75 -89.5 -89.25 ... 89.5 89.75 90.0
* longitude (longitude) float64 0.0 0.25 0.5 0.75 ... 359.0 359.2 359.5 359.8fig, ax = plt.subplots(
nrows=1, ncols=1,
figsize=(3, 1.5),
subplot_kw={'projection': ccrs.Robinson(central_longitude=210)},
constrained_layout=True)
# Plot model results using contourf
p0 = ax.contourf(tp.longitude, tp.latitude, tp,
levels=np.linspace(0, 16, 17),
cmap='YlGnBu', extend='both',
transform=ccrs.PlateCarree())
plt.colorbar(p0, ax=ax)
ax.coastlines(linewidth=0.5)
<cartopy.mpl.feature_artist.FeatureArtist at 0x1479a0bd03d0>
4.5. Example 3: Plot AMOC from TraCE and compare with McManus et al. 2004#
TraCE data is directly accessible on NCAR machines:
/glade/campaign/cesm/collections/TraCE
file = '/glade/campaign/cesm/collections/TraCE/ocn/proc/tavg/decadal/trace.01-36.22000BP.pop.MOC.22000BP_decavg_400BCE.nc'
ds = xr.open_dataset(file)
ds
<xarray.Dataset>
Dimensions: (nlat: 116, nlon: 100, time: 2204, transport_reg: 2,
moc_comp: 1, moc_z: 26, lat_aux_grid: 105, z_t: 25,
z_w: 25, transport_comp: 3)
Coordinates:
TLAT (nlat, nlon) float32 ...
TLONG (nlat, nlon) float32 ...
ULAT (nlat, nlon) float32 ...
ULONG (nlat, nlon) float32 ...
* lat_aux_grid (lat_aux_grid) float32 -80.26 -78.73 ... 88.38 90.0
moc_components (moc_comp) |S256 ...
* moc_z (moc_z) float32 0.0 800.0 ... 4.503e+05 5e+05
* time (time) float64 -22.0 -21.99 -21.98 ... 0.01 0.02 0.03
transport_regions (transport_reg) |S256 ...
* z_t (z_t) float32 400.0 1.222e+03 ... 4.255e+05 4.751e+05
* z_w (z_w) float32 0.0 800.0 ... 4.007e+05 4.503e+05
Dimensions without coordinates: nlat, nlon, transport_reg, moc_comp,
transport_comp
Data variables: (12/51)
ANGLE (nlat, nlon) float32 ...
ANGLET (nlat, nlon) float32 ...
DXT (nlat, nlon) float32 ...
DXU (nlat, nlon) float32 ...
DYT (nlat, nlon) float32 ...
DYU (nlat, nlon) float32 ...
... ...
sea_ice_salinity float64 ...
sflux_factor float64 ...
sound float64 ...
stefan_boltzmann float64 ...
transport_components (transport_comp) |S256 ...
vonkar float64 ...
Attributes:
title: b30.22_0kaDVT
contents: Diagnostic and Prognostic Variables
source: POP, the NCAR/CSM Ocean Component
revision: $Name: ccsm3_0_1_beta22 $
calendar: All years have exactly 365 days.
conventions: CF-1.0; http://www.cgd.ucar.edu/cms/eaton/netc...
start_time: This dataset was created on 2007-07-29 at 23:1...
cell_methods: cell_methods = time: mean ==> the variable val...
history: Fri Nov 8 22:51:40 2013: /glade/apps/opt/nco/...
nco_openmp_thread_number: 1
NCO: 4.3.24.5.1. Meridional Overturning Circulation is MOC, a 5-dimentional variable#
transport_componentshas three components: (1) Total, (2) Eulerian-Mean Advection, and (3) Eddy-Induced Advection (bolus) + Diffusionmoc_compofMOCis 0, which means the MOC caculation is the (1) total transporttransport_regionshas two parts: (1) Global Ocean - Marginal Seas and (2) Atlantic Ocean + Labrador Sea + GIN Sea + Arctic Oceantransport_regofMOCare 0 and 1, which indicate MOC values for Global Ocean and the Atlantic Ocean, respectivelyNote that the depth,
moc_z, is in centimeter
ds.MOC
<xarray.DataArray 'MOC' (time: 2204, transport_reg: 2, moc_comp: 1, moc_z: 26,
lat_aux_grid: 105)>
[12033840 values with dtype=float32]
Coordinates:
* lat_aux_grid (lat_aux_grid) float32 -80.26 -78.73 ... 88.38 90.0
moc_components (moc_comp) |S256 ...
* moc_z (moc_z) float32 0.0 800.0 1.644e+03 ... 4.503e+05 5e+05
* time (time) float64 -22.0 -21.99 -21.98 ... 0.01 0.02 0.03
transport_regions (transport_reg) |S256 ...
Dimensions without coordinates: transport_reg, moc_comp
Attributes:
long_name: Meridional Overturning Circulation
units: Sverdrupsds.transport_regions.values
array([b'Global Ocean - Marginal Seas',
b'Atlantic Ocean + Labrador Sea + GIN Sea + Arctic Ocean'],
dtype='|S256')
ds.MOC.transport_reg.values
array([0, 1])
ds.transport_components.values
array([b'Total', b'Eulerian-Mean Advection',
b'Eddy-Induced Advection (bolus) + Diffusion'], dtype='|S256')
ds.MOC.moc_comp.values
array([0])
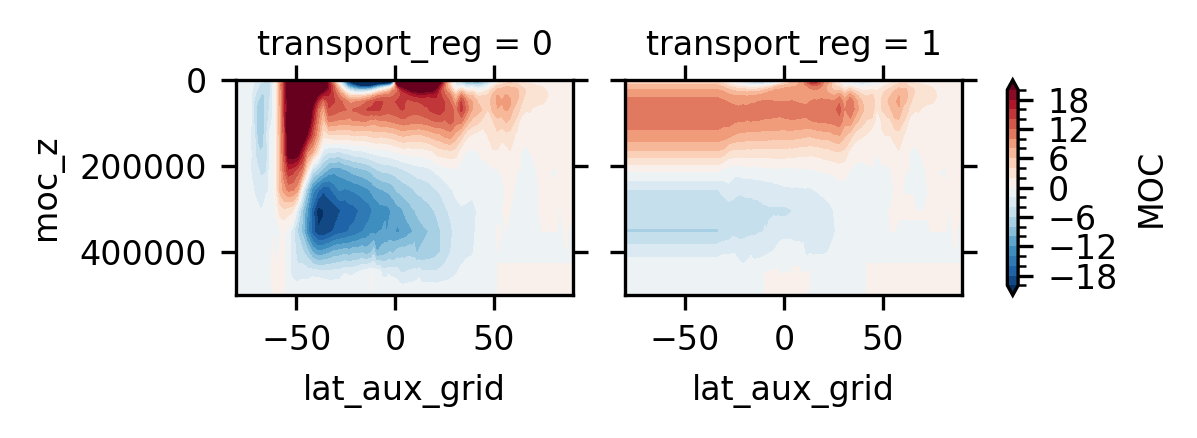
4.5.2. The left and right plots are the global mean and Atlantic MOC, respectively#
ds.MOC.isel(time=slice(0, 10), moc_comp=0).mean('time').plot.contourf(
size=1.5, x='lat_aux_grid', y='moc_z', col='transport_reg',
levels=np.linspace(-20, 20, 21))
plt.gca().invert_yaxis()
4.5.3. Again, transport_reg=1 means Atlantic and moc_comp=0 means the total transport#
amoc = ds.MOC.isel(transport_reg=1, moc_comp=0)
amoc
<xarray.DataArray 'MOC' (time: 2204, moc_z: 26, lat_aux_grid: 105)>
[6016920 values with dtype=float32]
Coordinates:
* lat_aux_grid (lat_aux_grid) float32 -80.26 -78.73 ... 88.38 90.0
moc_components |S256 ...
* moc_z (moc_z) float32 0.0 800.0 1.644e+03 ... 4.503e+05 5e+05
* time (time) float64 -22.0 -21.99 -21.98 ... 0.01 0.02 0.03
transport_regions |S256 b'Atlantic Ocean + Labrador Sea + GIN Sea + Arct...
Attributes:
long_name: Meridional Overturning Circulation
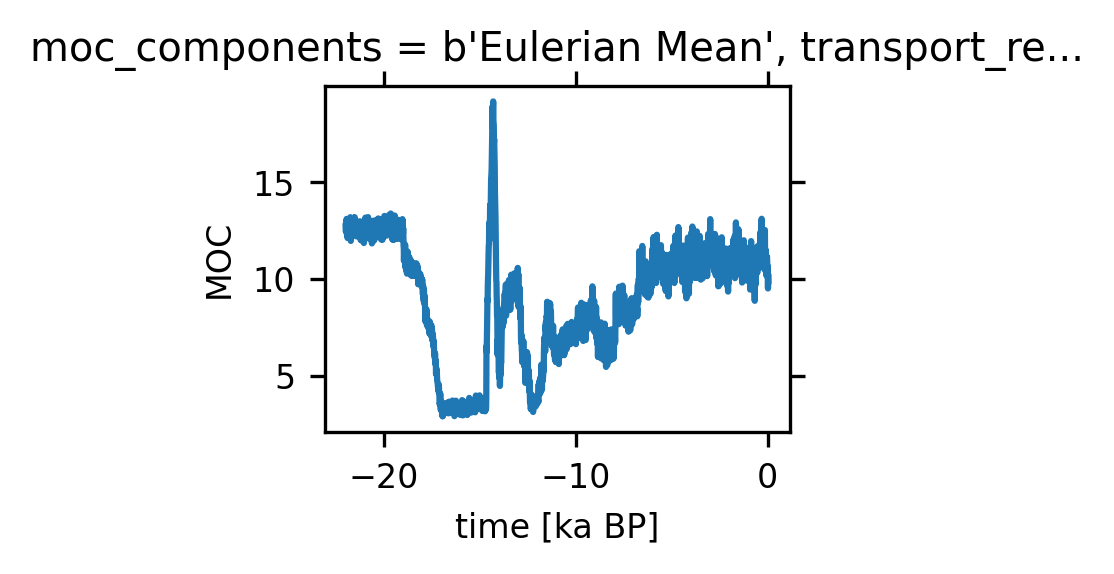
units: Sverdrups4.5.4. We define AMOC time series as the maximum over the North Atlantic and beneath a depth of 500 m (to avoid the wind-drive cell)#
amoc_ts = amoc.sel(moc_z=slice(50000, 500000),
lat_aux_grid=slice(0, 90)).max(('moc_z', 'lat_aux_grid'))
amoc_ts.plot(size=1.5)
[<matplotlib.lines.Line2D at 0x147993801750>]
4.5.5. Load the McManus et al. (2004) data from NOAA#
McManus04 = 'https://www.ncei.noaa.gov/pub/data/paleo/paleocean/sediment_files/complete/o326-gc5-tab.txt'
pa_th = pd.read_table(McManus04, header=37)
# Replace missing values
pa_th = pa_th[pa_th["pa/th232"] != -999].reset_index(drop=True)
pa_th.head()
| depth | calyrBP | pa/th232 | pa/th232.err | pa/th238 | pa/th238.err | d18Og.infla | Unnamed: 7 | |
|---|---|---|---|---|---|---|---|---|
| 0 | -999 | 100 | 0.054 | 0.002 | 0.055 | 0.002 | -999.00 | NaN |
| 1 | -999 | 480 | 0.054 | 0.002 | 0.054 | 0.002 | -999.00 | NaN |
| 2 | -999 | 960 | 0.057 | 0.004 | 0.057 | 0.004 | 0.66 | NaN |
| 3 | -999 | 1430 | 0.054 | 0.002 | 0.054 | 0.002 | -999.00 | NaN |
| 4 | -999 | 1910 | 0.055 | 0.002 | 0.055 | 0.002 | 0.63 | NaN |
# Change the time to be consistent with TraCE
pa_th['time'] = pa_th['calyrBP '] / -1000.0
pa_th.head()
| depth | calyrBP | pa/th232 | pa/th232.err | pa/th238 | pa/th238.err | d18Og.infla | Unnamed: 7 | time | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | -999 | 100 | 0.054 | 0.002 | 0.055 | 0.002 | -999.00 | NaN | -0.10 |
| 1 | -999 | 480 | 0.054 | 0.002 | 0.054 | 0.002 | -999.00 | NaN | -0.48 |
| 2 | -999 | 960 | 0.057 | 0.004 | 0.057 | 0.004 | 0.66 | NaN | -0.96 |
| 3 | -999 | 1430 | 0.054 | 0.002 | 0.054 | 0.002 | -999.00 | NaN | -1.43 |
| 4 | -999 | 1910 | 0.055 | 0.002 | 0.055 | 0.002 | 0.63 | NaN | -1.91 |
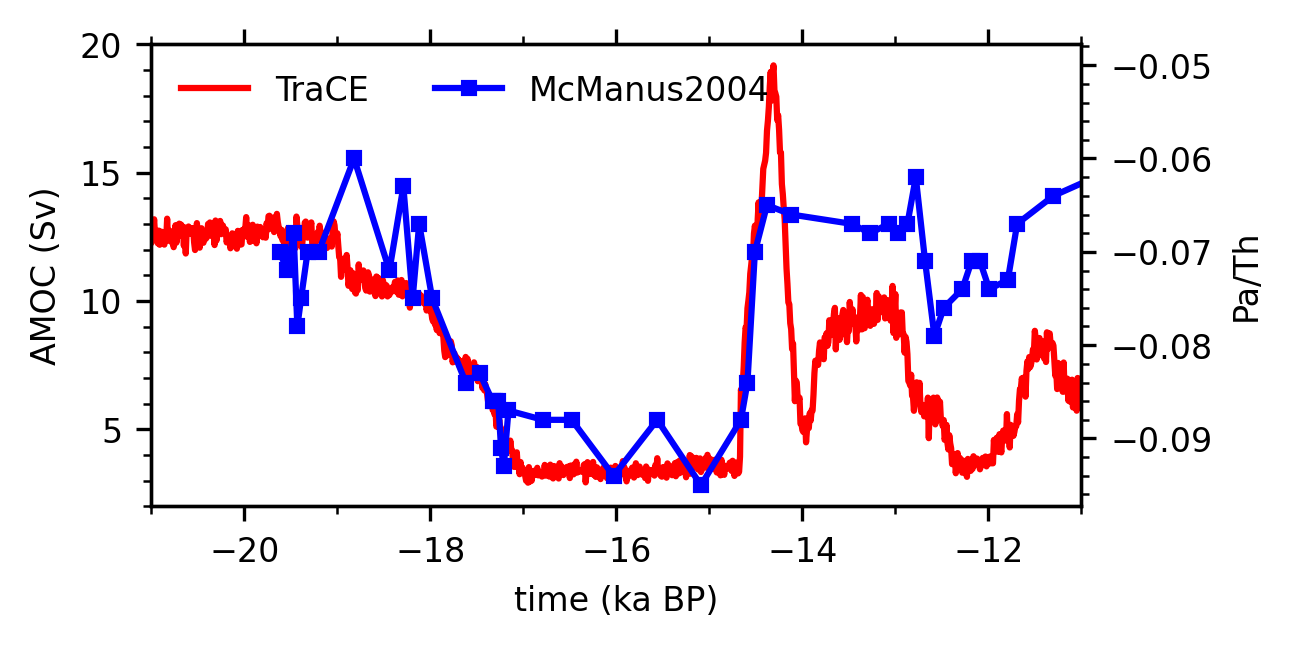
4.5.6. Plot time series#
fig, ax = plt.subplots(figsize=(4, 2))
# Plot the AMOC time series in TraCE
ax.plot(amoc_ts.time, amoc_ts, 'red',
label='TraCE')
ax.set_xlim([-21, -11])
ax.set_ylim([2, 20])
ax.set_xlabel('time (ka BP)')
ax.set_ylabel('AMOC (Sv)')
ax.xaxis.set_major_locator(plt.MultipleLocator(2))
ax.xaxis.set_minor_locator(plt.MultipleLocator(1))
ax.yaxis.set_major_locator(plt.MultipleLocator(5))
ax.yaxis.set_minor_locator(plt.MultipleLocator(1))
# Plot the Pa/Th time series in McManus et al. (2004)
ax2 = ax.twinx()
ax2.plot(pa_th['time'], pa_th['pa/th238']*-1,
'blue',
marker='s', markersize=3,
label='McManus2004')
ax2.set_ylabel('Pa/Th')
ax2.yaxis.set_major_locator(plt.MultipleLocator(0.01))
ax2.yaxis.set_minor_locator(plt.MultipleLocator(0.002))
# Add a legend
lh1, ll1 = ax.get_legend_handles_labels()
lh2, ll2 = ax2.get_legend_handles_labels()
leg = ax.legend(lh1+lh2, ll1+ll2, frameon=False,
loc='upper left', ncol=2, fontsize=8)
4.6. Example 4. Compute and plot precipitation δ18O#
We define a function to compute precipitation δ18O
def calculate_d18Op(ds):
"""
Compute precipitation δ18O with iCESM output
Parameters
ds: xarray.Dataset contains necessary variables
Returns
ds: xarray.Dataset with δ18O added
"""
# convective & large-scale rain and snow, respectively
p16O = ds.PRECRC_H216Or + ds.PRECSC_H216Os + ds.PRECRL_H216OR + ds.PRECSL_H216OS
p18O = ds.PRECRC_H218Or + ds.PRECSC_H218Os + ds.PRECRL_H218OR + ds.PRECSL_H218OS
# avoid dividing by small number here
p18O = p18O.where(p16O > 1.E-18, 1.E-18)
p16O = p16O.where(p16O > 1.E-18, 1.E-18)
d18O = (p18O / p16O - 1.0) * 1000.0
ds['p16O'] = p16O
ds['p18O'] = p18O
ds['d18O'] = d18O
return ds
Figure out the file names
!ls /glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.iPI.01/atm/proc/tseries/month_1/*PRECRC_H218Or*
!ls /glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.i21ka.03/atm/proc/tseries/month_1/*PRECRC_H218Or*
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.iPI.01/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.iPI.01.cam.h0.PRECRC_H218Or.0001-0900.cal_adj.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.iPI.01/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.iPI.01.cam.h0.PRECRC_H218Or.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.i21ka.03/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.i21ka.03.cam.h0.PRECRC_H218Or.0001-0900.cal_adj.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.i21ka.03/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.i21ka.03.cam.h0.PRECRC_H218Or.0001-0900.nc
We need to read in all these variables
vnames = ['PRECC', 'PRECL',
'PRECRC_H216Or', 'PRECSC_H216Os', 'PRECRL_H216OR', 'PRECSL_H216OS',
'PRECRC_H218Or', 'PRECSC_H218Os', 'PRECRL_H218OR', 'PRECSL_H218OS']
storage_dir = '/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/'
hist_dir = '/atm/proc/tseries/month_1/'
case_pre = 'b.e12.B1850C5.f19_g16.iPI.01'
case_lgm = 'b.e12.B1850C5.f19_g16.i21ka.03'
fnames_pre = []
fnames_lgm = []
for vname in vnames:
fname = glob.glob(storage_dir + case_pre + hist_dir + '*' + vname + '.0001-0900.nc')
fnames_pre.extend(fname)
fname = glob.glob(storage_dir + case_lgm + hist_dir + '*' + vname + '.0001-0900.nc')
fnames_lgm.extend(fname)
print(*fnames_pre, sep='\n')
print(*fnames_lgm, sep='\n')
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.iPI.01/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.iPI.01.cam.h0.PRECC.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.iPI.01/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.iPI.01.cam.h0.PRECL.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.iPI.01/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.iPI.01.cam.h0.PRECRC_H216Or.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.iPI.01/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.iPI.01.cam.h0.PRECSC_H216Os.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.iPI.01/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.iPI.01.cam.h0.PRECRL_H216OR.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.iPI.01/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.iPI.01.cam.h0.PRECSL_H216OS.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.iPI.01/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.iPI.01.cam.h0.PRECRC_H218Or.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.iPI.01/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.iPI.01.cam.h0.PRECSC_H218Os.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.iPI.01/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.iPI.01.cam.h0.PRECRL_H218OR.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.iPI.01/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.iPI.01.cam.h0.PRECSL_H218OS.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.i21ka.03/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.i21ka.03.cam.h0.PRECC.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.i21ka.03/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.i21ka.03.cam.h0.PRECL.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.i21ka.03/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.i21ka.03.cam.h0.PRECRC_H216Or.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.i21ka.03/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.i21ka.03.cam.h0.PRECSC_H216Os.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.i21ka.03/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.i21ka.03.cam.h0.PRECRL_H216OR.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.i21ka.03/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.i21ka.03.cam.h0.PRECSL_H216OS.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.i21ka.03/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.i21ka.03.cam.h0.PRECRC_H218Or.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.i21ka.03/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.i21ka.03.cam.h0.PRECSC_H218Os.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.i21ka.03/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.i21ka.03.cam.h0.PRECRL_H218OR.0001-0900.nc
/glade/campaign/cgd/ppc/jiangzhu/iCESM1.2/b.e12.B1850C5.f19_g16.i21ka.03/atm/proc/tseries/month_1/b.e12.B1850C5.f19_g16.i21ka.03.cam.h0.PRECSL_H218OS.0001-0900.nc
%%time
ds_pre = xr.open_mfdataset(fnames_pre, parallel=True,
data_vars='minimal',
coords='minimal',
compat='override',
chunks={'time':12}).isel(time=slice(-120, None))
ds_lgm = xr.open_mfdataset(fnames_lgm, parallel=True,
data_vars='minimal',
coords='minimal',
compat='override',
chunks={'time':12}).isel(time=slice(-120, None))
CPU times: user 1.33 s, sys: 184 ms, total: 1.51 s
Wall time: 5.13 s
ds_pre = calculate_d18Op(ds_pre)
ds_lgm = calculate_d18Op(ds_lgm)
ds_pre
<xarray.Dataset>
Dimensions: (lev: 30, ilev: 31, lat: 96, lon: 144, slat: 95, slon: 144,
time: 120, nbnd: 2)
Coordinates:
* lev (lev) float64 3.643 7.595 14.36 24.61 ... 957.5 976.3 992.6
* ilev (ilev) float64 2.255 5.032 10.16 18.56 ... 967.5 985.1 1e+03
* lat (lat) float64 -90.0 -88.11 -86.21 -84.32 ... 86.21 88.11 90.0
* lon (lon) float64 0.0 2.5 5.0 7.5 ... 350.0 352.5 355.0 357.5
* slat (slat) float64 -89.05 -87.16 -85.26 ... 85.26 87.16 89.05
* slon (slon) float64 -1.25 1.25 3.75 6.25 ... 351.2 353.8 356.2
* time (time) object 0891-02-01 00:00:00 ... 0901-01-01 00:00:00
Dimensions without coordinates: nbnd
Data variables: (12/44)
hyam (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
hybm (lev) float64 dask.array<chunksize=(30,), meta=np.ndarray>
hyai (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
hybi (ilev) float64 dask.array<chunksize=(31,), meta=np.ndarray>
P0 float64 ...
w_stag (slat) float64 dask.array<chunksize=(95,), meta=np.ndarray>
... ...
PRECSC_H218Os (time, lat, lon) float32 dask.array<chunksize=(12, 96, 144), meta=np.ndarray>
PRECSL_H216OS (time, lat, lon) float32 dask.array<chunksize=(12, 96, 144), meta=np.ndarray>
PRECSL_H218OS (time, lat, lon) float32 dask.array<chunksize=(12, 96, 144), meta=np.ndarray>
p16O (time, lat, lon) float32 dask.array<chunksize=(12, 96, 144), meta=np.ndarray>
p18O (time, lat, lon) float32 dask.array<chunksize=(12, 96, 144), meta=np.ndarray>
d18O (time, lat, lon) float32 dask.array<chunksize=(12, 96, 144), meta=np.ndarray>
Attributes:
Conventions: CF-1.0
source: CAM
case: b.e12.B1850C5.f19_g16.iPI.01
title: UNSET
logname: jiangzhu
host: r5i1n15
Version: $Name$
revision_Id: $Id$
initial_file: b.ie12.B1850C5CN.f19_g16.09.cam.i.0401-01-01-00000.nc
topography_file: /glade/p/cesmdata/cseg/inputdata/atm/cam/topo/USGS-gtop...# Save netcdf data
# ds_pre.to_netcdf('dataset_with_d18Op.nc')
d18Op_pre = ds_pre.d18O.mean('time')
d18Op_lgm = ds_lgm.d18O.mean('time')
# Sometimes, it is preferred to use the precipitation-amount weighted d18O
#d18Op_pre = ds_pre.d18O.weighted(ds_pre.p16O).mean('time')
d18Op_pre
<xarray.DataArray 'd18O' (lat: 96, lon: 144)> dask.array<mean_agg-aggregate, shape=(96, 144), dtype=float32, chunksize=(96, 144), chunktype=numpy.ndarray> Coordinates: * lat (lat) float64 -90.0 -88.11 -86.21 -84.32 ... 84.32 86.21 88.11 90.0 * lon (lon) float64 0.0 2.5 5.0 7.5 10.0 ... 350.0 352.5 355.0 357.5
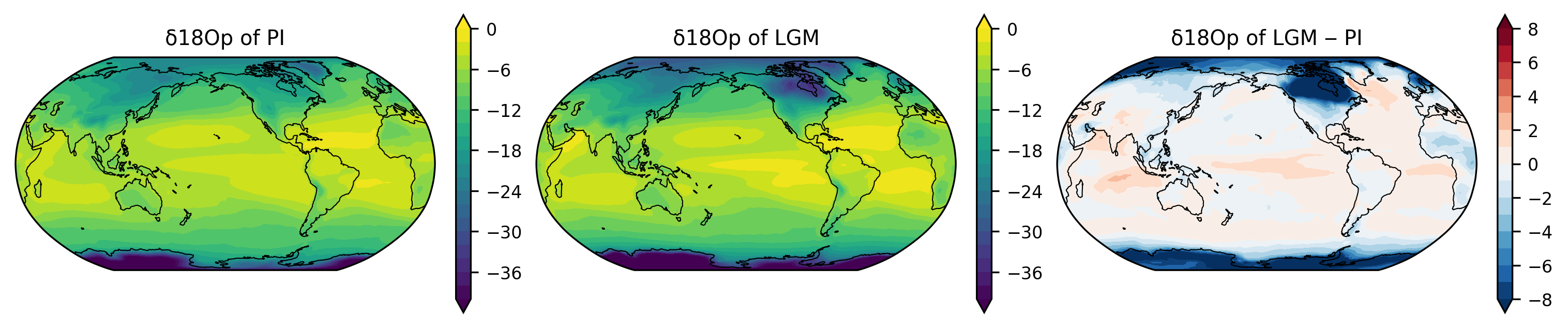
fig, axes = plt.subplots(1, 3,
figsize=(10, 2),
subplot_kw={'projection': ccrs.Robinson(central_longitude=210)},
constrained_layout=True)
axes = axes.ravel()
lon = d18Op_pre.lon
lat = d18Op_pre.lat
ax = axes[0]
var_new, lon_new = add_cyclic_point(d18Op_pre, lon)
p0 = ax.contourf(lon_new, lat, var_new,
levels=np.linspace(-40, 0, 21),
extend='both',
transform=ccrs.PlateCarree())
plt.colorbar(p0, ax=ax)
ax.set_title("δ18Op of PI")
ax = axes[1]
var_new, lon_new = add_cyclic_point(d18Op_lgm, lon)
p1 = ax.contourf(lon_new, lat, var_new,
levels=np.linspace(-40, 0, 21),
extend='both',
transform=ccrs.PlateCarree())
plt.colorbar(p1, ax=ax)
ax.set_title("δ18Op of LGM")
ax = axes[2]
var_diff = d18Op_lgm - d18Op_pre
var_new, lon_new = add_cyclic_point(var_diff, lon)
p2 = ax.contourf(lon_new, lat, var_new,
cmap='RdBu_r',
levels=np.linspace(-8, 8, 17),
extend='both',
transform=ccrs.PlateCarree())
plt.colorbar(p2, ax=ax)
ax.set_title("δ18Op of LGM ‒ PI")
for ax in axes:
ax.set_global()
ax.coastlines(linewidth=0.5)