
0.2 Python & Jupyter Basics#
Tutorials at the 2025 paleoCAMP | June 16–June 30, 2025
Jiang Zhu
jiangzhu@ucar.edu
Climate & Global Dynamics Laboratory
NSF National Center for Atmospheric Research
Why Python?#
Python is open source.
Python is a high-productivity language: do more with less code.
Python is interpreted and portable, like Matlab, so no need to compile your code for it to run.
Python is highly human readable and popular. Currently there are 600,000+ Python packages on the Python Package Index (PyPI).
Pangeo is the recommended Python software stack:#
Xarray: Read and write NetCDF files
Jupyter: A browser-based, “terminal”-like interface with nice graphics capabilities
Additional packages provide useful capabilities, such as:
numpy: For general Python N-dimensional arrays
matplotlib: For general plotting capabilities
cartopy: For plotting data on map projections
pandas: For importing/exporting files in common formats (e.g., csv, xlsx) and organize and manipulate the data in a handy object
scipy: For scientific computing (e.g., optimization, linear algebra, statistics, file I/O for formats like .mat) based on numpy arrays
hvplot: A high-level plotting API for the PyData ecosystem built on HoloViews.
xesmf: Xarray-based regridding package
scikit-learn: For data mining and data analysis
“How to install external Python libs on my laptop / desktop?”#
To best way to get the scientific Python environment is using the Conda package management system. Please follow the official installation guide for installing on
Hello world#
print("Hello world!")
Hello world!
Comparison between Python, C, and Matlab#
Python uses 0 based ordering
Python uses
#sign for commentingPython determines block size based on indentation, i.e., no
endor}to close a block

Figure: Python vs C vs Matlab
Jupyter#
Jupyter is a web server for interactive computing that supports multiple programming languages.
When you run it on your laptop (Jupyter Notebook or JupyterLab), you can directly interact with the web server from your browser.
When working remotely (e.g., on a HPC), you just communicate HTML (and serialized data) back and forth from your browser to the web server.
Reproducibility & Extensibility#
It is fast and efficient.
Jupyter allows you to save your work in a notebook (ipynb), so you can share your work with other people simply by sharing your notebook file. Other people can use it and extend it for their own purposes.
A Jupyter Notebook is composed of cells. A Cell could have three types#
Codecell contains Python codeMarkdowncell has Markup language and can be used for notes/commentsRawcell has simple text
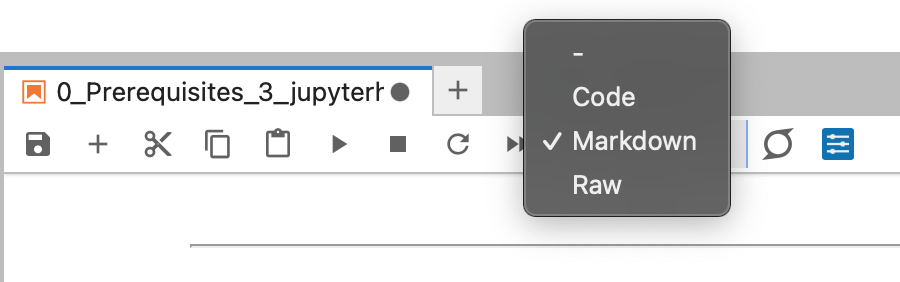
Figure: Cell types
How to run a cell?#
To run a cell in a notebook. Click on the cell and press: Shift+return

Figure: Running a cell
How to run a shell command within a cell?#
# list the content of within the current directory
!ls
0.1_demo_unix.ipynb 7_run_challenging_LGM.ipynb
0.2_intro_python_jupyter.ipynb _build
0.3_demo_ncar_account_jupyterhub.ipynb _config.yml
1_intro_to_CESM.ipynb images
2_demo_run_CESM_4steps.ipynb intro.md
3_analyze_CESM_output.ipynb LICENSE
4a_opt1_run_piControl_midHolocene.ipynb precip_xy.era5.png
4b_opt2_analyze_long_midHolocene.ipynb README.md
5_info_Data_Access.ipynb _toc.yml
6_info_iCESM_iTRACE.ipynb
%ls
0.1_demo_unix.ipynb 7_run_challenging_LGM.ipynb
0.2_intro_python_jupyter.ipynb _build/
0.3_demo_ncar_account_jupyterhub.ipynb _config.yml
1_intro_to_CESM.ipynb images/
2_demo_run_CESM_4steps.ipynb intro.md
3_analyze_CESM_output.ipynb LICENSE
4a_opt1_run_piControl_midHolocene.ipynb precip_xy.era5.png
4b_opt2_analyze_long_midHolocene.ipynb README.md
5_info_Data_Access.ipynb _toc.yml
6_info_iCESM_iTRACE.ipynb
