
3. Analyze CESM Output#
Tutorials at the 2025 paleoCAMP | June 16–June 30, 2025
Jiang Zhu
jiangzhu@ucar.edu
Climate & Global Dynamics Laboratory
NSF National Center for Atmospheric Research
Learning Objectives:
Learn to use the NCAR JupyterHub for data access and analysis
Learn to read and examine netcdf files using Xarray
Learn techniques to make basic visualization of temperature, precipitation, and sea-surface temperature
Time to learn: 60 minutes
How to get started?
Launch a JupyterHub server: https://jupyterhub.hpc.ucar.edu/
Sign in with your
usernameandpassword,passcodepasswordis the CIT password you set up with CISLpasscodeis the six digits on your DUO appNOTE the comma
,betweenpasswordandpasscode
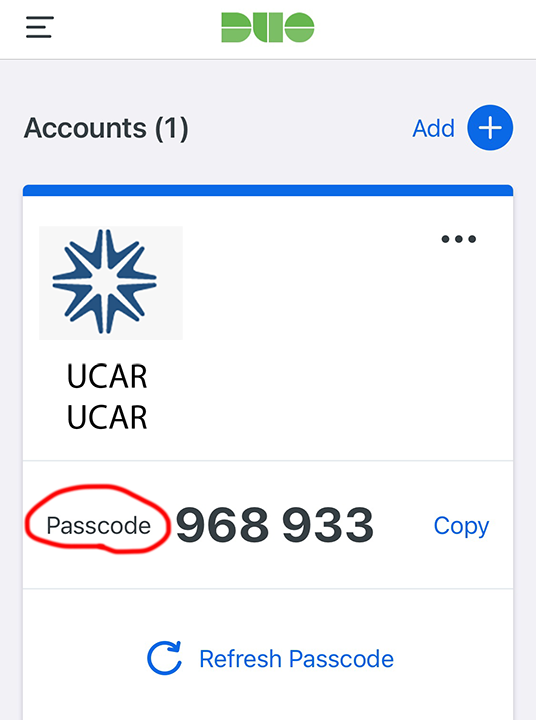
Figure: Screenshot of DUO app
Start the JupyterHub. From the dropdown menu, select
Casper PBS Batchfor Resource Selectioncasperfor Queue or ReservationUAZN0042for Project Account06:00:00for Wall Time
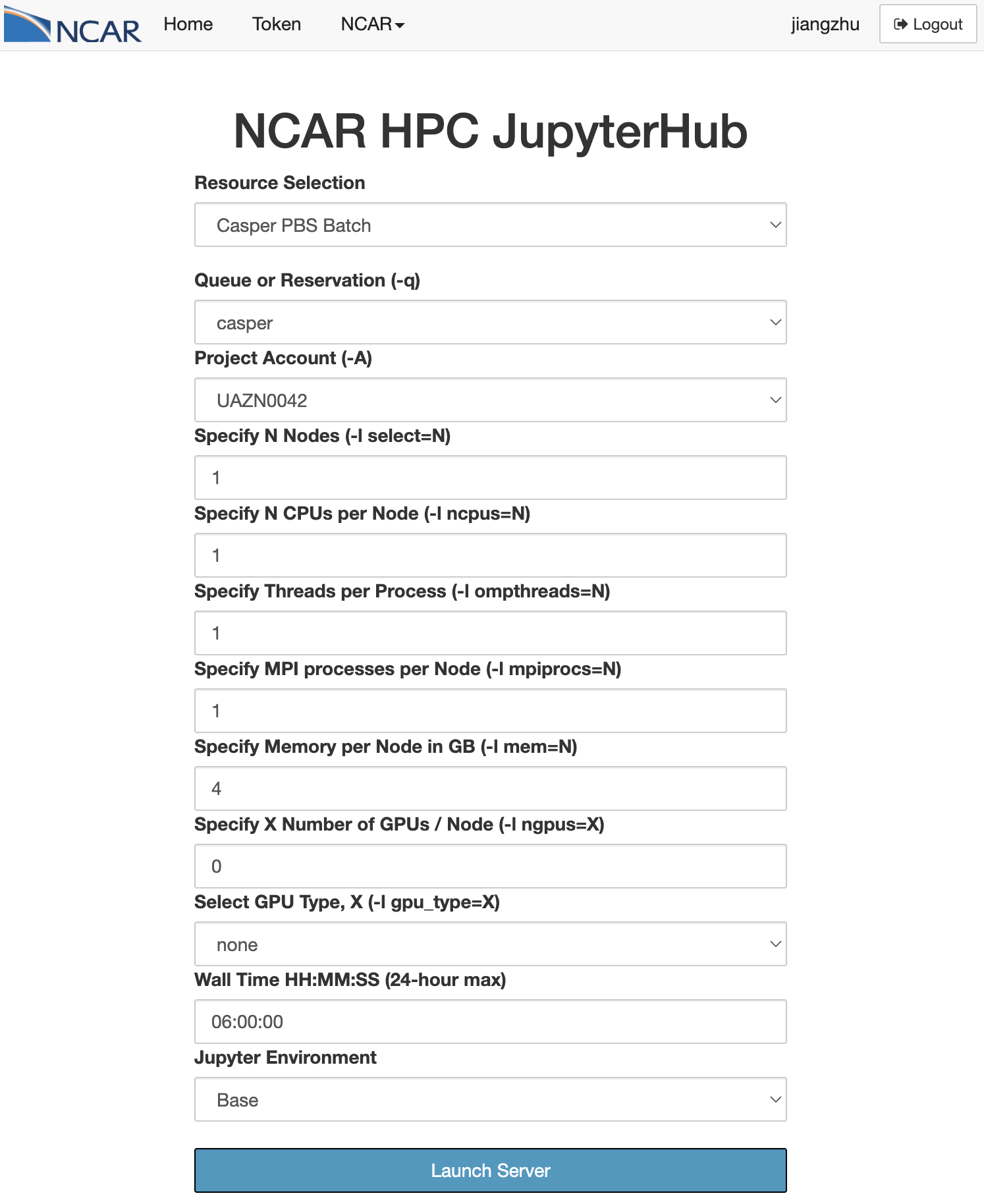
Figure: Jupyterhub Casper
Lauch a terminal: click
File,New, andTerminal
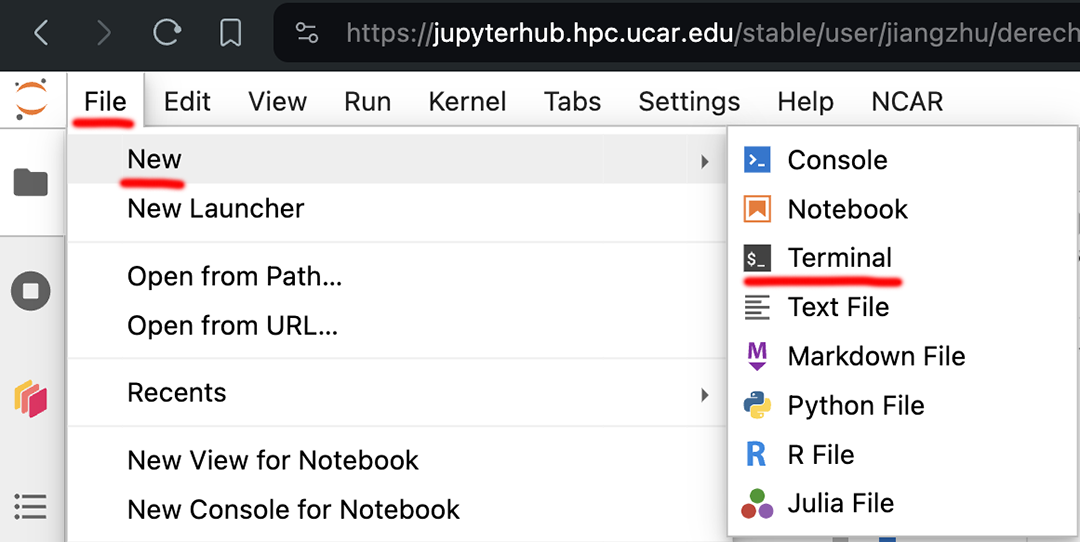
Figure: Launch a Terminal
Type
git clone https://github.com/jiang-zhu/paleocamp2025.gitFind and open
3_analyze_CESM_output.ipynbfrom the left sidebarMake sure to use
NPL 2025aas your Kernel (select from top right corner)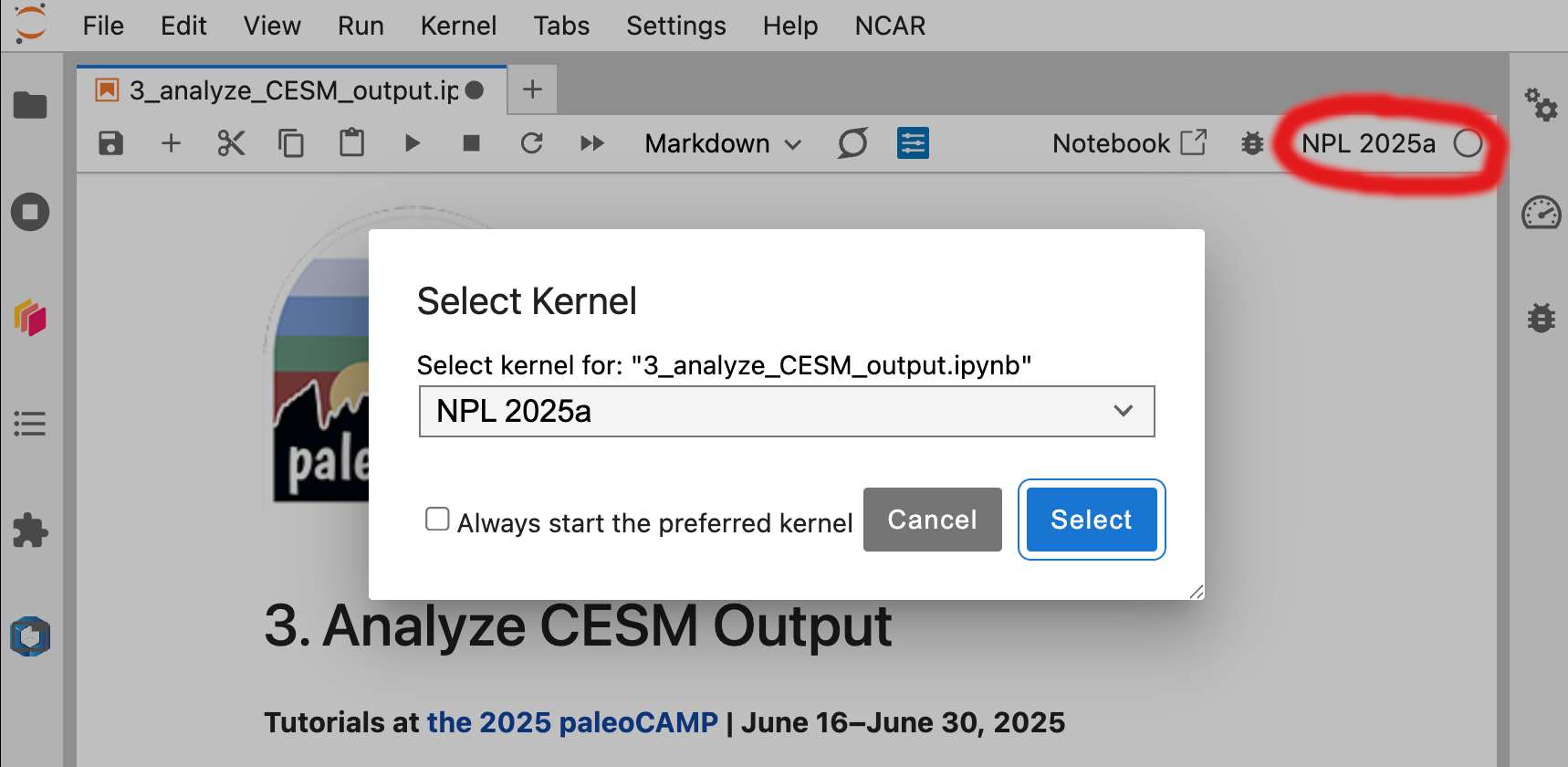
Figure: Launch a Terminal
Load Python packages
import os
import glob
from datetime import timedelta
import xarray as xr
import numpy as np
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
from cartopy.util import add_cyclic_point
# geocat is used for interpolation of the atmosphere output
from geocat.comp import interpolation
# xesmf is used for regridding the ocean output
import xesmf
# hvplot provides interactive plots
import hvplot.xarray
Analysis 1: plot solar insolation in the MH to further validate the simulation#
Load data#
I have both the piControl and midHolocene simulations finished.
I see 12-month history files for each simulation in my
Archive Directory.
!ls /glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/*cam.h0.0001*
!ls /glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/*cam.h0.0001*
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-01.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-02.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-03.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-04.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-05.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-06.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-07.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-08.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-09.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-10.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-11.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-12.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-01.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-02.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-03.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-04.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-05.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-06.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-07.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-08.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-09.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-10.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-11.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-12.nc
Use
globandos.path.jointo obtain the file names including path.Use wildcard
*to catch all files in the atmosphere history directory (see Wildcard in 0_1_quick_demo_unix.ipynb).
storage_dir = '/glade/derecho/scratch/jiangzhu/archive/'
hist_dir = 'atm/hist'
case_PI = 'b.e21.B1850.f19_g17.piControl.001'
case_MH = 'b.e21.B1850.f19_g17.midHolocene.001'
files_PI = glob.glob(os.path.join(storage_dir, case_PI, hist_dir, '*.cam.h0.0001*'))
files_MH = glob.glob(os.path.join(storage_dir, case_MH, hist_dir, '*.cam.h0.0001*'))
print('List of files for PI')
print(*files_PI, sep='\n')
print('List of files for MH')
print(*files_MH, sep='\n')
List of files for PI
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-10.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-02.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-01.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-11.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-06.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-03.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-05.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-12.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-04.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-09.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-07.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/atm/hist/b.e21.B1850.f19_g17.piControl.001.cam.h0.0001-08.nc
List of files for MH
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-04.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-06.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-07.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-03.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-11.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-08.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-01.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-05.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-02.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-09.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-12.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/atm/hist/b.e21.B1850.f19_g17.midHolocene.001.cam.h0.0001-10.nc
Use
xr.open_mfdatasetto open multiple files at once in parallel (12 month in this case)We will keep using the Xarray Datasets
ds_MHandds_PIthroughout the Notebook
ds_PI = xr.open_mfdataset(files_PI)
ds_MH = xr.open_mfdataset(files_MH)
ds_PI
<xarray.Dataset> Size: 541MB
Dimensions: (time: 12, zlon: 1, nbnd: 2, lat: 96, lev: 26, ilev: 27,
lon: 144)
Coordinates:
* lat (lat) float64 768B -90.0 -88.11 -86.21 ... 86.21 88.11 90.0
* zlon (zlon) float64 8B 0.0
* lon (lon) float64 1kB 0.0 2.5 5.0 7.5 ... 350.0 352.5 355.0 357.5
* lev (lev) float64 208B 3.545 7.389 13.97 ... 929.6 970.6 992.6
* ilev (ilev) float64 216B 2.194 4.895 9.882 ... 956.0 985.1 1e+03
* time (time) object 96B 0001-02-01 00:00:00 ... 0002-01-01 00:00:00
Dimensions without coordinates: nbnd
Data variables: (12/122)
zlon_bnds (time, zlon, nbnd) float64 192B dask.array<chunksize=(1, 1, 2), meta=np.ndarray>
gw (time, lat) float64 9kB dask.array<chunksize=(1, 96), meta=np.ndarray>
hyam (time, lev) float64 2kB dask.array<chunksize=(1, 26), meta=np.ndarray>
hybm (time, lev) float64 2kB dask.array<chunksize=(1, 26), meta=np.ndarray>
P0 (time) float64 96B 1e+05 1e+05 1e+05 ... 1e+05 1e+05 1e+05
hyai (time, ilev) float64 3kB dask.array<chunksize=(1, 27), meta=np.ndarray>
... ...
VU (time, lev, lat, lon) float32 17MB dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
VV (time, lev, lat, lon) float32 17MB dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
Vzm (time, ilev, lat, zlon) float32 124kB dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
WTHzm (time, ilev, lat, zlon) float32 124kB dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
Wzm (time, ilev, lat, zlon) float32 124kB dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
Z3 (time, lev, lat, lon) float32 17MB dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
Attributes:
Conventions: CF-1.0
source: CAM
case: b.e21.B1850.f19_g17.piControl.001
logname: jiangzhu
host: derecho2
initial_file: /glade/campaign/cesm/cesmdata/inputdata/atm/cam/inic/f...
topography_file: /glade/campaign/cesm/cesmdata/inputdata/atm/cam/topo/f...
model_doi_url: https://doi.org/10.5065/D67H1H0V
time_period_freq: month_1- time: 12
- zlon: 1
- nbnd: 2
- lat: 96
- lev: 26
- ilev: 27
- lon: 144
- lat(lat)float64-90.0 -88.11 -86.21 ... 88.11 90.0
- long_name :
- latitude
- units :
- degrees_north
array([-90. , -88.105263, -86.210526, -84.315789, -82.421053, -80.526316, -78.631579, -76.736842, -74.842105, -72.947368, -71.052632, -69.157895, -67.263158, -65.368421, -63.473684, -61.578947, -59.684211, -57.789474, -55.894737, -54. , -52.105263, -50.210526, -48.315789, -46.421053, -44.526316, -42.631579, -40.736842, -38.842105, -36.947368, -35.052632, -33.157895, -31.263158, -29.368421, -27.473684, -25.578947, -23.684211, -21.789474, -19.894737, -18. , -16.105263, -14.210526, -12.315789, -10.421053, -8.526316, -6.631579, -4.736842, -2.842105, -0.947368, 0.947368, 2.842105, 4.736842, 6.631579, 8.526316, 10.421053, 12.315789, 14.210526, 16.105263, 18. , 19.894737, 21.789474, 23.684211, 25.578947, 27.473684, 29.368421, 31.263158, 33.157895, 35.052632, 36.947368, 38.842105, 40.736842, 42.631579, 44.526316, 46.421053, 48.315789, 50.210526, 52.105263, 54. , 55.894737, 57.789474, 59.684211, 61.578947, 63.473684, 65.368421, 67.263158, 69.157895, 71.052632, 72.947368, 74.842105, 76.736842, 78.631579, 80.526316, 82.421053, 84.315789, 86.210526, 88.105263, 90. ]) - zlon(zlon)float640.0
- long_name :
- longitude
- units :
- degrees_east
- bounds :
- zlon_bnds
array([0.])
- lon(lon)float640.0 2.5 5.0 ... 352.5 355.0 357.5
- long_name :
- longitude
- units :
- degrees_east
array([ 0. , 2.5, 5. , 7.5, 10. , 12.5, 15. , 17.5, 20. , 22.5, 25. , 27.5, 30. , 32.5, 35. , 37.5, 40. , 42.5, 45. , 47.5, 50. , 52.5, 55. , 57.5, 60. , 62.5, 65. , 67.5, 70. , 72.5, 75. , 77.5, 80. , 82.5, 85. , 87.5, 90. , 92.5, 95. , 97.5, 100. , 102.5, 105. , 107.5, 110. , 112.5, 115. , 117.5, 120. , 122.5, 125. , 127.5, 130. , 132.5, 135. , 137.5, 140. , 142.5, 145. , 147.5, 150. , 152.5, 155. , 157.5, 160. , 162.5, 165. , 167.5, 170. , 172.5, 175. , 177.5, 180. , 182.5, 185. , 187.5, 190. , 192.5, 195. , 197.5, 200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. , 332.5, 335. , 337.5, 340. , 342.5, 345. , 347.5, 350. , 352.5, 355. , 357.5]) - lev(lev)float643.545 7.389 13.97 ... 970.6 992.6
- long_name :
- hybrid level at midpoints (1000*(A+B))
- units :
- hPa
- positive :
- down
- standard_name :
- atmosphere_hybrid_sigma_pressure_coordinate
- formula_terms :
- a: hyam b: hybm p0: P0 ps: PS
array([ 3.544638, 7.388814, 13.967214, 23.944625, 37.23029 , 53.114605, 70.05915 , 85.439115, 100.514695, 118.250335, 139.115395, 163.66207 , 192.539935, 226.513265, 266.481155, 313.501265, 368.81798 , 433.895225, 510.455255, 600.5242 , 696.79629 , 787.70206 , 867.16076 , 929.648875, 970.55483 , 992.5561 ]) - ilev(ilev)float642.194 4.895 9.882 ... 985.1 1e+03
- long_name :
- hybrid level at interfaces (1000*(A+B))
- units :
- hPa
- positive :
- down
- standard_name :
- atmosphere_hybrid_sigma_pressure_coordinate
- formula_terms :
- a: hyai b: hybi p0: P0 ps: PS
array([ 2.194067, 4.895209, 9.882418, 18.05201 , 29.83724 , 44.62334 , 61.60587 , 78.51243 , 92.3658 , 108.66359 , 127.83708 , 150.39371 , 176.93043 , 208.14944 , 244.87709 , 288.08522 , 338.91731 , 398.71865 , 469.0718 , 551.83871 , 649.20969 , 744.38289 , 831.02123 , 903.30029 , 955.99746 , 985.1122 , 1000. ]) - time(time)object0001-02-01 00:00:00 ... 0002-01-...
- long_name :
- time
- bounds :
- time_bnds
array([cftime.DatetimeNoLeap(1, 2, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 3, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 4, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 5, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 6, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 7, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 8, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 9, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 10, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 11, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 12, 1, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(2, 1, 1, 0, 0, 0, 0, has_year_zero=True)], dtype=object)
- zlon_bnds(time, zlon, nbnd)float64dask.array<chunksize=(1, 1, 2), meta=np.ndarray>
- long_name :
- zlon bounds
- units :
- degrees_east
Array Chunk Bytes 192 B 16 B Shape (12, 1, 2) (1, 1, 2) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - gw(time, lat)float64dask.array<chunksize=(1, 96), meta=np.ndarray>
- long_name :
- latitude weights
Array Chunk Bytes 9.00 kiB 768 B Shape (12, 96) (1, 96) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - hyam(time, lev)float64dask.array<chunksize=(1, 26), meta=np.ndarray>
- long_name :
- hybrid A coefficient at layer midpoints
Array Chunk Bytes 2.44 kiB 208 B Shape (12, 26) (1, 26) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - hybm(time, lev)float64dask.array<chunksize=(1, 26), meta=np.ndarray>
- long_name :
- hybrid B coefficient at layer midpoints
Array Chunk Bytes 2.44 kiB 208 B Shape (12, 26) (1, 26) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - P0(time)float641e+05 1e+05 1e+05 ... 1e+05 1e+05
- long_name :
- reference pressure
- units :
- Pa
array([100000., 100000., 100000., 100000., 100000., 100000., 100000., 100000., 100000., 100000., 100000., 100000.]) - hyai(time, ilev)float64dask.array<chunksize=(1, 27), meta=np.ndarray>
- long_name :
- hybrid A coefficient at layer interfaces
Array Chunk Bytes 2.53 kiB 216 B Shape (12, 27) (1, 27) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - hybi(time, ilev)float64dask.array<chunksize=(1, 27), meta=np.ndarray>
- long_name :
- hybrid B coefficient at layer interfaces
Array Chunk Bytes 2.53 kiB 216 B Shape (12, 27) (1, 27) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - date(time)int32dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- current date (YYYYMMDD)
Array Chunk Bytes 48 B 4 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type int32 numpy.ndarray - datesec(time)int32dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- current seconds of current date
Array Chunk Bytes 48 B 4 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type int32 numpy.ndarray - time_bnds(time, nbnd)objectdask.array<chunksize=(1, 2), meta=np.ndarray>
- long_name :
- time interval endpoints
Array Chunk Bytes 192 B 16 B Shape (12, 2) (1, 2) Dask graph 12 chunks in 25 graph layers Data type object numpy.ndarray - date_written(time)|S8dask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 37 graph layers Data type |S8 numpy.ndarray - time_written(time)|S8dask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 37 graph layers Data type |S8 numpy.ndarray - ndbase(time)int320 0 0 0 0 0 0 0 0 0 0 0
- long_name :
- base day
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], dtype=int32)
- nsbase(time)int320 0 0 0 0 0 0 0 0 0 0 0
- long_name :
- seconds of base day
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], dtype=int32)
- nbdate(time)int3210101 10101 10101 ... 10101 10101
- long_name :
- base date (YYYYMMDD)
array([10101, 10101, 10101, 10101, 10101, 10101, 10101, 10101, 10101, 10101, 10101, 10101], dtype=int32) - nbsec(time)int320 0 0 0 0 0 0 0 0 0 0 0
- long_name :
- seconds of base date
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], dtype=int32)
- mdt(time)int321800 1800 1800 ... 1800 1800 1800
- long_name :
- timestep
- units :
- s
array([1800, 1800, 1800, 1800, 1800, 1800, 1800, 1800, 1800, 1800, 1800, 1800], dtype=int32) - ndcur(time)int32dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- current day (from base day)
Array Chunk Bytes 48 B 4 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type int32 numpy.ndarray - nscur(time)int32dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- current seconds of current day
Array Chunk Bytes 48 B 4 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type int32 numpy.ndarray - co2vmr(time)float64dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- co2 volume mixing ratio
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type float64 numpy.ndarray - ch4vmr(time)float64dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- ch4 volume mixing ratio
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type float64 numpy.ndarray - n2ovmr(time)float64dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- n2o volume mixing ratio
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type float64 numpy.ndarray - f11vmr(time)float64dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- f11 volume mixing ratio
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type float64 numpy.ndarray - f12vmr(time)float64dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- f12 volume mixing ratio
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type float64 numpy.ndarray - sol_tsi(time)float64dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- total solar irradiance
- units :
- W/m2
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type float64 numpy.ndarray - nsteph(time)int32dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- current timestep
Array Chunk Bytes 48 B 4 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type int32 numpy.ndarray - AEROD_v(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- 1
- long_name :
- Total Aerosol Optical Depth in visible band
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CLDHGH(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- fraction
- long_name :
- Vertically-integrated high cloud
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CLDICE(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- kg/kg
- mixing_ratio :
- wet
- long_name :
- Grid box averaged cloud ice amount
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CLDLIQ(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- kg/kg
- mixing_ratio :
- wet
- long_name :
- Grid box averaged cloud liquid amount
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CLDLOW(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- fraction
- long_name :
- Vertically-integrated low cloud
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CLDMED(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- fraction
- long_name :
- Vertically-integrated mid-level cloud
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CLDTOT(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- fraction
- long_name :
- Vertically-integrated total cloud
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CLOUD(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- Sampling_Sequence :
- rad_lwsw
- units :
- fraction
- long_name :
- Cloud fraction
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CONCLD(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- fraction
- long_name :
- Convective cloud cover
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - DCQ(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- kg/kg/s
- long_name :
- Q tendency due to moist processes
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - DTCOND(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- K/s
- long_name :
- T tendency - moist processes
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - DTV(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- K/s
- long_name :
- T vertical diffusion
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - EMIS(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- cloud emissivity
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FICE(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- fraction
- long_name :
- Fractional ice content within cloud
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLDS(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Downwelling longwave flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLDSC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky downwelling longwave flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLNS(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net longwave flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLNSC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky net longwave flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLNT(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net longwave flux at top of model
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLNTC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky net longwave flux at top of model
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLUT(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Upwelling longwave flux at top of model
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLUTC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky upwelling longwave flux at top of model
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSDS(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Downwelling solar flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSDSC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky downwelling solar flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSDTOA(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Downwelling solar flux at top of atmosphere
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSNS(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net solar flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSNSC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky net solar flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSNT(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net solar flux at top of model
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSNTC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky net solar flux at top of model
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSNTOA(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net solar flux at top of atmosphere
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSNTOAC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky net solar flux at top of atmosphere
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSUTOA(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Upwelling solar flux at top of atmosphere
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ICEFRAC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- fraction
- long_name :
- Fraction of sfc area covered by sea-ice
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ICIMR(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- kg/kg
- long_name :
- Prognostic in-cloud ice mixing ratio
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ICWMR(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- kg/kg
- long_name :
- Prognostic in-cloud water mixing ratio
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - LANDFRAC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- fraction
- long_name :
- Fraction of sfc area covered by land
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - LHFLX(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- W/m2
- long_name :
- Surface latent heat flux
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - LWCF(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Longwave cloud forcing
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - MSKtem(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- 1
- long_name :
- TEM mask
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - OCNFRAC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- fraction
- long_name :
- Fraction of sfc area covered by ocean
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - OMEGA(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- Pa/s
- long_name :
- Vertical velocity (pressure)
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - OMEGAT(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- K Pa/s
- long_name :
- Vertical heat flux
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PBLH(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m
- long_name :
- PBL height
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PHIS(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m2/s2
- long_name :
- Surface geopotential
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PRECC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m/s
- long_name :
- Convective precipitation rate (liq + ice)
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PRECL(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m/s
- long_name :
- Large-scale (stable) precipitation rate (liq + ice)
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PRECSC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m/s
- long_name :
- Convective snow rate (water equivalent)
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PRECSL(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m/s
- long_name :
- Large-scale (stable) snow rate (water equivalent)
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PS(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- Pa
- long_name :
- Surface pressure
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PSL(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- Pa
- long_name :
- Sea level pressure
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - Q(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- kg/kg
- mixing_ratio :
- wet
- long_name :
- Specific humidity
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - QFLX(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- kg/m2/s
- long_name :
- Surface water flux
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - QREFHT(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- kg/kg
- long_name :
- Reference height humidity
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - QRL(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- Sampling_Sequence :
- rad_lwsw
- units :
- K/s
- long_name :
- Longwave heating rate
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - QRS(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- Sampling_Sequence :
- rad_lwsw
- units :
- K/s
- long_name :
- Solar heating rate
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - RELHUM(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- percent
- long_name :
- Relative humidity
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SFCLDICE(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- kg/m2/s
- long_name :
- CLDICE surface flux
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SFCLDLIQ(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- kg/m2/s
- long_name :
- CLDLIQ surface flux
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SHFLX(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- W/m2
- long_name :
- Surface sensible heat flux
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SNOWHICE(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m
- long_name :
- Snow depth over ice
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SNOWHLND(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m
- long_name :
- Water equivalent snow depth
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SOLIN(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Solar insolation
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SWCF(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Shortwave cloud forcing
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - T(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- K
- long_name :
- Temperature
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TAUGWX(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- N/m2
- long_name :
- Zonal gravity wave surface stress
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TAUGWY(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- N/m2
- long_name :
- Meridional gravity wave surface stress
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TAUX(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- N/m2
- long_name :
- Zonal surface stress
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TAUY(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- N/m2
- long_name :
- Meridional surface stress
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TGCLDCWP(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- gram/m2
- long_name :
- Total grid-box cloud water path (liquid and ice)
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TGCLDIWP(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- gram/m2
- long_name :
- Total grid-box cloud ice water path
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TGCLDLWP(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- gram/m2
- long_name :
- Total grid-box cloud liquid water path
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TH(time, ilev, lat, lon)float32dask.array<chunksize=(1, 27, 96, 144), meta=np.ndarray>
- mdims :
- 2
- units :
- K
- long_name :
- Potential Temperature
- cell_methods :
- time: mean
Array Chunk Bytes 17.09 MiB 1.42 MiB Shape (12, 27, 96, 144) (1, 27, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - THzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- K
- long_name :
- Zonal-Mean potential temp - defined on ilev
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TMQ(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- kg/m2
- long_name :
- Total (vertically integrated) precipitable water
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TREFHT(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- K
- long_name :
- Reference height temperature
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TS(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- K
- long_name :
- Surface temperature (radiative)
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TSMN(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- K
- long_name :
- Minimum surface temperature over output period
- cell_methods :
- time: minimum
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TSMX(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- K
- long_name :
- Maximum surface temperature over output period
- cell_methods :
- time: maximum
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - U(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m/s
- long_name :
- Zonal wind
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - U10(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m/s
- long_name :
- 10m wind speed
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - UTGWORO(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m/s2
- long_name :
- U tendency - orographic gravity wave drag
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - UU(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m2/s2
- long_name :
- Zonal velocity squared
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - UVzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- M2/S2
- long_name :
- Meridional Flux of Zonal Momentum: 3D zon. mean
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - UWzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- M2/S2
- long_name :
- Vertical Flux of Zonal Momentum: 3D zon. mean
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - Uzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- M/S
- long_name :
- Zonal-Mean zonal wind - defined on ilev
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - V(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m/s
- long_name :
- Meridional wind
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VD01(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- kg/kg/s
- long_name :
- Vertical diffusion of Q
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VQ(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m/skg/kg
- long_name :
- Meridional water transport
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VT(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- K m/s
- long_name :
- Meridional heat transport
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VTHzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- MK/S
- long_name :
- Meridional Heat Flux: 3D zon. mean
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VU(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m2/s2
- long_name :
- Meridional flux of zonal momentum
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VV(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m2/s2
- long_name :
- Meridional velocity squared
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - Vzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- M/S
- long_name :
- Zonal-Mean meridional wind - defined on ilev
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - WTHzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- MK/S
- long_name :
- Vertical Heat Flux: 3D zon. mean
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - Wzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- M/S
- long_name :
- Zonal-Mean vertical wind - defined on ilev
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - Z3(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m
- long_name :
- Geopotential Height (above sea level)
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray
- latPandasIndex
PandasIndex(Index([ -90.0, -88.10526315789474, -86.21052631578948, -84.3157894736842, -82.42105263157895, -80.52631578947368, -78.63157894736842, -76.73684210526316, -74.84210526315789, -72.94736842105263, -71.05263157894737, -69.15789473684211, -67.26315789473685, -65.36842105263158, -63.473684210526315, -61.578947368421055, -59.684210526315795, -57.78947368421053, -55.89473684210527, -54.0, -52.10526315789474, -50.21052631578947, -48.31578947368421, -46.42105263157895, -44.526315789473685, -42.631578947368425, -40.73684210526316, -38.8421052631579, -36.94736842105264, -35.05263157894737, -33.15789473684211, -31.263157894736842, -29.368421052631582, -27.473684210526322, -25.578947368421055, -23.684210526315795, -21.789473684210535, -19.89473684210526, -18.0, -16.10526315789474, -14.21052631578948, -12.31578947368422, -10.421052631578945, -8.526315789473685, -6.631578947368425, -4.736842105263165, -2.8421052631579045, -0.9473684210526301, 0.9473684210526301, 2.8421052631578902, 4.73684210526315, 6.631578947368411, 8.526315789473685, 10.421052631578945, 12.315789473684205, 14.210526315789465, 16.105263157894726, 18.0, 19.89473684210526, 21.78947368421052, 23.68421052631578, 25.57894736842104, 27.473684210526315, 29.368421052631575, 31.263157894736835, 33.157894736842096, 35.052631578947356, 36.94736842105263, 38.84210526315789, 40.73684210526315, 42.63157894736841, 44.52631578947367, 46.42105263157893, 48.31578947368419, 50.21052631578948, 52.10526315789474, 54.0, 55.89473684210526, 57.78947368421052, 59.68421052631578, 61.57894736842104, 63.4736842105263, 65.36842105263156, 67.26315789473682, 69.15789473684211, 71.05263157894737, 72.94736842105263, 74.84210526315789, 76.73684210526315, 78.63157894736841, 80.52631578947367, 82.42105263157893, 84.31578947368419, 86.21052631578945, 88.10526315789474, 90.0], dtype='float64', name='lat')) - zlonPandasIndex
PandasIndex(Index([0.0], dtype='float64', name='zlon'))
- lonPandasIndex
PandasIndex(Index([ 0.0, 2.5, 5.0, 7.5, 10.0, 12.5, 15.0, 17.5, 20.0, 22.5, ... 335.0, 337.5, 340.0, 342.5, 345.0, 347.5, 350.0, 352.5, 355.0, 357.5], dtype='float64', name='lon', length=144)) - levPandasIndex
PandasIndex(Index([3.5446380000000097, 7.3888135000000075, 13.967214000000006, 23.944625, 37.23029000000011, 53.1146050000002, 70.05915000000029, 85.43911500000031, 100.51469500000029, 118.25033500000026, 139.11539500000046, 163.66207000000043, 192.53993500000033, 226.51326500000036, 266.4811550000001, 313.5012650000006, 368.81798000000157, 433.8952250000011, 510.45525500000167, 600.5242000000027, 696.7962900000033, 787.7020600000026, 867.1607600000013, 929.6488750000024, 970.5548300000014, 992.5560999999998], dtype='float64', name='lev')) - ilevPandasIndex
PandasIndex(Index([ 2.194067000000008, 4.895209000000012, 9.882418000000005, 18.052010000000006, 29.837239999999987, 44.62334000000023, 61.60587000000017, 78.5124300000004, 92.36580000000022, 108.66359000000037, 127.83708000000016, 150.3937100000008, 176.9304300000001, 208.14944000000057, 244.87709000000012, 288.08522000000016, 338.9173100000011, 398.71865000000196, 469.0718000000003, 551.8387100000031, 649.2096900000024, 744.3828900000042, 831.0212300000011, 903.3002900000016, 955.9974600000032, 985.1121999999997, 1000.0], dtype='float64', name='ilev')) - timePandasIndex
PandasIndex(CFTimeIndex([0001-02-01 00:00:00, 0001-03-01 00:00:00, 0001-04-01 00:00:00, 0001-05-01 00:00:00, 0001-06-01 00:00:00, 0001-07-01 00:00:00, 0001-08-01 00:00:00, 0001-09-01 00:00:00, 0001-10-01 00:00:00, 0001-11-01 00:00:00, 0001-12-01 00:00:00, 0002-01-01 00:00:00], dtype='object', length=12, calendar='noleap', freq='MS'))
- Conventions :
- CF-1.0
- source :
- CAM
- case :
- b.e21.B1850.f19_g17.piControl.001
- logname :
- jiangzhu
- host :
- derecho2
- initial_file :
- /glade/campaign/cesm/cesmdata/inputdata/atm/cam/inic/fv/cami_0000-01-01_1.9x2.5_L26_c070408.nc
- topography_file :
- /glade/campaign/cesm/cesmdata/inputdata/atm/cam/topo/fv_1.9x2.5_nc3000_Nsw084_Nrs016_Co120_Fi001_ZR_GRNL_031819.nc
- model_doi_url :
- https://doi.org/10.5065/D67H1H0V
- time_period_freq :
- month_1
# We need this fix to get the correct time, i.e., January to December of year 1
ds_PI['time'] = ds_PI.time.get_index('time') - timedelta(days=15)
ds_MH['time'] = ds_MH.time.get_index('time') - timedelta(days=15)
ds_PI
# Explore the dataset and find the Solar insolation, SOLIN
<xarray.Dataset> Size: 541MB
Dimensions: (time: 12, zlon: 1, nbnd: 2, lat: 96, lev: 26, ilev: 27,
lon: 144)
Coordinates:
* lat (lat) float64 768B -90.0 -88.11 -86.21 ... 86.21 88.11 90.0
* zlon (zlon) float64 8B 0.0
* lon (lon) float64 1kB 0.0 2.5 5.0 7.5 ... 350.0 352.5 355.0 357.5
* lev (lev) float64 208B 3.545 7.389 13.97 ... 929.6 970.6 992.6
* ilev (ilev) float64 216B 2.194 4.895 9.882 ... 956.0 985.1 1e+03
* time (time) object 96B 0001-01-17 00:00:00 ... 0001-12-17 00:00:00
Dimensions without coordinates: nbnd
Data variables: (12/122)
zlon_bnds (time, zlon, nbnd) float64 192B dask.array<chunksize=(1, 1, 2), meta=np.ndarray>
gw (time, lat) float64 9kB dask.array<chunksize=(1, 96), meta=np.ndarray>
hyam (time, lev) float64 2kB dask.array<chunksize=(1, 26), meta=np.ndarray>
hybm (time, lev) float64 2kB dask.array<chunksize=(1, 26), meta=np.ndarray>
P0 (time) float64 96B 1e+05 1e+05 1e+05 ... 1e+05 1e+05 1e+05
hyai (time, ilev) float64 3kB dask.array<chunksize=(1, 27), meta=np.ndarray>
... ...
VU (time, lev, lat, lon) float32 17MB dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
VV (time, lev, lat, lon) float32 17MB dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
Vzm (time, ilev, lat, zlon) float32 124kB dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
WTHzm (time, ilev, lat, zlon) float32 124kB dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
Wzm (time, ilev, lat, zlon) float32 124kB dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
Z3 (time, lev, lat, lon) float32 17MB dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
Attributes:
Conventions: CF-1.0
source: CAM
case: b.e21.B1850.f19_g17.piControl.001
logname: jiangzhu
host: derecho2
initial_file: /glade/campaign/cesm/cesmdata/inputdata/atm/cam/inic/f...
topography_file: /glade/campaign/cesm/cesmdata/inputdata/atm/cam/topo/f...
model_doi_url: https://doi.org/10.5065/D67H1H0V
time_period_freq: month_1- time: 12
- zlon: 1
- nbnd: 2
- lat: 96
- lev: 26
- ilev: 27
- lon: 144
- lat(lat)float64-90.0 -88.11 -86.21 ... 88.11 90.0
- long_name :
- latitude
- units :
- degrees_north
array([-90. , -88.105263, -86.210526, -84.315789, -82.421053, -80.526316, -78.631579, -76.736842, -74.842105, -72.947368, -71.052632, -69.157895, -67.263158, -65.368421, -63.473684, -61.578947, -59.684211, -57.789474, -55.894737, -54. , -52.105263, -50.210526, -48.315789, -46.421053, -44.526316, -42.631579, -40.736842, -38.842105, -36.947368, -35.052632, -33.157895, -31.263158, -29.368421, -27.473684, -25.578947, -23.684211, -21.789474, -19.894737, -18. , -16.105263, -14.210526, -12.315789, -10.421053, -8.526316, -6.631579, -4.736842, -2.842105, -0.947368, 0.947368, 2.842105, 4.736842, 6.631579, 8.526316, 10.421053, 12.315789, 14.210526, 16.105263, 18. , 19.894737, 21.789474, 23.684211, 25.578947, 27.473684, 29.368421, 31.263158, 33.157895, 35.052632, 36.947368, 38.842105, 40.736842, 42.631579, 44.526316, 46.421053, 48.315789, 50.210526, 52.105263, 54. , 55.894737, 57.789474, 59.684211, 61.578947, 63.473684, 65.368421, 67.263158, 69.157895, 71.052632, 72.947368, 74.842105, 76.736842, 78.631579, 80.526316, 82.421053, 84.315789, 86.210526, 88.105263, 90. ]) - zlon(zlon)float640.0
- long_name :
- longitude
- units :
- degrees_east
- bounds :
- zlon_bnds
array([0.])
- lon(lon)float640.0 2.5 5.0 ... 352.5 355.0 357.5
- long_name :
- longitude
- units :
- degrees_east
array([ 0. , 2.5, 5. , 7.5, 10. , 12.5, 15. , 17.5, 20. , 22.5, 25. , 27.5, 30. , 32.5, 35. , 37.5, 40. , 42.5, 45. , 47.5, 50. , 52.5, 55. , 57.5, 60. , 62.5, 65. , 67.5, 70. , 72.5, 75. , 77.5, 80. , 82.5, 85. , 87.5, 90. , 92.5, 95. , 97.5, 100. , 102.5, 105. , 107.5, 110. , 112.5, 115. , 117.5, 120. , 122.5, 125. , 127.5, 130. , 132.5, 135. , 137.5, 140. , 142.5, 145. , 147.5, 150. , 152.5, 155. , 157.5, 160. , 162.5, 165. , 167.5, 170. , 172.5, 175. , 177.5, 180. , 182.5, 185. , 187.5, 190. , 192.5, 195. , 197.5, 200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. , 332.5, 335. , 337.5, 340. , 342.5, 345. , 347.5, 350. , 352.5, 355. , 357.5]) - lev(lev)float643.545 7.389 13.97 ... 970.6 992.6
- long_name :
- hybrid level at midpoints (1000*(A+B))
- units :
- hPa
- positive :
- down
- standard_name :
- atmosphere_hybrid_sigma_pressure_coordinate
- formula_terms :
- a: hyam b: hybm p0: P0 ps: PS
array([ 3.544638, 7.388814, 13.967214, 23.944625, 37.23029 , 53.114605, 70.05915 , 85.439115, 100.514695, 118.250335, 139.115395, 163.66207 , 192.539935, 226.513265, 266.481155, 313.501265, 368.81798 , 433.895225, 510.455255, 600.5242 , 696.79629 , 787.70206 , 867.16076 , 929.648875, 970.55483 , 992.5561 ]) - ilev(ilev)float642.194 4.895 9.882 ... 985.1 1e+03
- long_name :
- hybrid level at interfaces (1000*(A+B))
- units :
- hPa
- positive :
- down
- standard_name :
- atmosphere_hybrid_sigma_pressure_coordinate
- formula_terms :
- a: hyai b: hybi p0: P0 ps: PS
array([ 2.194067, 4.895209, 9.882418, 18.05201 , 29.83724 , 44.62334 , 61.60587 , 78.51243 , 92.3658 , 108.66359 , 127.83708 , 150.39371 , 176.93043 , 208.14944 , 244.87709 , 288.08522 , 338.91731 , 398.71865 , 469.0718 , 551.83871 , 649.20969 , 744.38289 , 831.02123 , 903.30029 , 955.99746 , 985.1122 , 1000. ]) - time(time)object0001-01-17 00:00:00 ... 0001-12-...
array([cftime.DatetimeNoLeap(1, 1, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 2, 14, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 3, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 4, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 5, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 6, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 7, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 8, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 9, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 10, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 12, 17, 0, 0, 0, 0, has_year_zero=True)], dtype=object)
- zlon_bnds(time, zlon, nbnd)float64dask.array<chunksize=(1, 1, 2), meta=np.ndarray>
- long_name :
- zlon bounds
- units :
- degrees_east
Array Chunk Bytes 192 B 16 B Shape (12, 1, 2) (1, 1, 2) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - gw(time, lat)float64dask.array<chunksize=(1, 96), meta=np.ndarray>
- long_name :
- latitude weights
Array Chunk Bytes 9.00 kiB 768 B Shape (12, 96) (1, 96) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - hyam(time, lev)float64dask.array<chunksize=(1, 26), meta=np.ndarray>
- long_name :
- hybrid A coefficient at layer midpoints
Array Chunk Bytes 2.44 kiB 208 B Shape (12, 26) (1, 26) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - hybm(time, lev)float64dask.array<chunksize=(1, 26), meta=np.ndarray>
- long_name :
- hybrid B coefficient at layer midpoints
Array Chunk Bytes 2.44 kiB 208 B Shape (12, 26) (1, 26) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - P0(time)float641e+05 1e+05 1e+05 ... 1e+05 1e+05
- long_name :
- reference pressure
- units :
- Pa
array([100000., 100000., 100000., 100000., 100000., 100000., 100000., 100000., 100000., 100000., 100000., 100000.]) - hyai(time, ilev)float64dask.array<chunksize=(1, 27), meta=np.ndarray>
- long_name :
- hybrid A coefficient at layer interfaces
Array Chunk Bytes 2.53 kiB 216 B Shape (12, 27) (1, 27) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - hybi(time, ilev)float64dask.array<chunksize=(1, 27), meta=np.ndarray>
- long_name :
- hybrid B coefficient at layer interfaces
Array Chunk Bytes 2.53 kiB 216 B Shape (12, 27) (1, 27) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - date(time)int32dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- current date (YYYYMMDD)
Array Chunk Bytes 48 B 4 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type int32 numpy.ndarray - datesec(time)int32dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- current seconds of current date
Array Chunk Bytes 48 B 4 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type int32 numpy.ndarray - time_bnds(time, nbnd)objectdask.array<chunksize=(1, 2), meta=np.ndarray>
- long_name :
- time interval endpoints
Array Chunk Bytes 192 B 16 B Shape (12, 2) (1, 2) Dask graph 12 chunks in 25 graph layers Data type object numpy.ndarray - date_written(time)|S8dask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 37 graph layers Data type |S8 numpy.ndarray - time_written(time)|S8dask.array<chunksize=(1,), meta=np.ndarray>
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 37 graph layers Data type |S8 numpy.ndarray - ndbase(time)int320 0 0 0 0 0 0 0 0 0 0 0
- long_name :
- base day
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], dtype=int32)
- nsbase(time)int320 0 0 0 0 0 0 0 0 0 0 0
- long_name :
- seconds of base day
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], dtype=int32)
- nbdate(time)int3210101 10101 10101 ... 10101 10101
- long_name :
- base date (YYYYMMDD)
array([10101, 10101, 10101, 10101, 10101, 10101, 10101, 10101, 10101, 10101, 10101, 10101], dtype=int32) - nbsec(time)int320 0 0 0 0 0 0 0 0 0 0 0
- long_name :
- seconds of base date
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0], dtype=int32)
- mdt(time)int321800 1800 1800 ... 1800 1800 1800
- long_name :
- timestep
- units :
- s
array([1800, 1800, 1800, 1800, 1800, 1800, 1800, 1800, 1800, 1800, 1800, 1800], dtype=int32) - ndcur(time)int32dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- current day (from base day)
Array Chunk Bytes 48 B 4 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type int32 numpy.ndarray - nscur(time)int32dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- current seconds of current day
Array Chunk Bytes 48 B 4 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type int32 numpy.ndarray - co2vmr(time)float64dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- co2 volume mixing ratio
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type float64 numpy.ndarray - ch4vmr(time)float64dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- ch4 volume mixing ratio
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type float64 numpy.ndarray - n2ovmr(time)float64dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- n2o volume mixing ratio
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type float64 numpy.ndarray - f11vmr(time)float64dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- f11 volume mixing ratio
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type float64 numpy.ndarray - f12vmr(time)float64dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- f12 volume mixing ratio
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type float64 numpy.ndarray - sol_tsi(time)float64dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- total solar irradiance
- units :
- W/m2
Array Chunk Bytes 96 B 8 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type float64 numpy.ndarray - nsteph(time)int32dask.array<chunksize=(1,), meta=np.ndarray>
- long_name :
- current timestep
Array Chunk Bytes 48 B 4 B Shape (12,) (1,) Dask graph 12 chunks in 25 graph layers Data type int32 numpy.ndarray - AEROD_v(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- 1
- long_name :
- Total Aerosol Optical Depth in visible band
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CLDHGH(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- fraction
- long_name :
- Vertically-integrated high cloud
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CLDICE(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- kg/kg
- mixing_ratio :
- wet
- long_name :
- Grid box averaged cloud ice amount
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CLDLIQ(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- kg/kg
- mixing_ratio :
- wet
- long_name :
- Grid box averaged cloud liquid amount
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CLDLOW(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- fraction
- long_name :
- Vertically-integrated low cloud
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CLDMED(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- fraction
- long_name :
- Vertically-integrated mid-level cloud
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CLDTOT(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- fraction
- long_name :
- Vertically-integrated total cloud
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CLOUD(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- Sampling_Sequence :
- rad_lwsw
- units :
- fraction
- long_name :
- Cloud fraction
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - CONCLD(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- fraction
- long_name :
- Convective cloud cover
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - DCQ(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- kg/kg/s
- long_name :
- Q tendency due to moist processes
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - DTCOND(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- K/s
- long_name :
- T tendency - moist processes
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - DTV(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- K/s
- long_name :
- T vertical diffusion
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - EMIS(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- Sampling_Sequence :
- rad_lwsw
- units :
- 1
- long_name :
- cloud emissivity
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FICE(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- fraction
- long_name :
- Fractional ice content within cloud
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLDS(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Downwelling longwave flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLDSC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky downwelling longwave flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLNS(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net longwave flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLNSC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky net longwave flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLNT(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net longwave flux at top of model
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLNTC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky net longwave flux at top of model
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLUT(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Upwelling longwave flux at top of model
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FLUTC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky upwelling longwave flux at top of model
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSDS(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Downwelling solar flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSDSC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky downwelling solar flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSDTOA(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Downwelling solar flux at top of atmosphere
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSNS(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net solar flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSNSC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky net solar flux at surface
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSNT(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net solar flux at top of model
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSNTC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky net solar flux at top of model
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSNTOA(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Net solar flux at top of atmosphere
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSNTOAC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Clearsky net solar flux at top of atmosphere
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FSUTOA(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Upwelling solar flux at top of atmosphere
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ICEFRAC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- fraction
- long_name :
- Fraction of sfc area covered by sea-ice
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ICIMR(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- kg/kg
- long_name :
- Prognostic in-cloud ice mixing ratio
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ICWMR(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- kg/kg
- long_name :
- Prognostic in-cloud water mixing ratio
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - LANDFRAC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- fraction
- long_name :
- Fraction of sfc area covered by land
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - LHFLX(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- W/m2
- long_name :
- Surface latent heat flux
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - LWCF(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Longwave cloud forcing
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - MSKtem(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- 1
- long_name :
- TEM mask
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - OCNFRAC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- fraction
- long_name :
- Fraction of sfc area covered by ocean
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - OMEGA(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- Pa/s
- long_name :
- Vertical velocity (pressure)
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - OMEGAT(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- K Pa/s
- long_name :
- Vertical heat flux
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PBLH(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m
- long_name :
- PBL height
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PHIS(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m2/s2
- long_name :
- Surface geopotential
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PRECC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m/s
- long_name :
- Convective precipitation rate (liq + ice)
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PRECL(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m/s
- long_name :
- Large-scale (stable) precipitation rate (liq + ice)
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PRECSC(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m/s
- long_name :
- Convective snow rate (water equivalent)
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PRECSL(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m/s
- long_name :
- Large-scale (stable) snow rate (water equivalent)
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PS(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- Pa
- long_name :
- Surface pressure
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PSL(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- Pa
- long_name :
- Sea level pressure
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - Q(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- kg/kg
- mixing_ratio :
- wet
- long_name :
- Specific humidity
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - QFLX(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- kg/m2/s
- long_name :
- Surface water flux
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - QREFHT(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- kg/kg
- long_name :
- Reference height humidity
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - QRL(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- Sampling_Sequence :
- rad_lwsw
- units :
- K/s
- long_name :
- Longwave heating rate
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - QRS(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- Sampling_Sequence :
- rad_lwsw
- units :
- K/s
- long_name :
- Solar heating rate
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - RELHUM(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- percent
- long_name :
- Relative humidity
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SFCLDICE(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- kg/m2/s
- long_name :
- CLDICE surface flux
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SFCLDLIQ(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- kg/m2/s
- long_name :
- CLDLIQ surface flux
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SHFLX(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- W/m2
- long_name :
- Surface sensible heat flux
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SNOWHICE(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m
- long_name :
- Snow depth over ice
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SNOWHLND(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m
- long_name :
- Water equivalent snow depth
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SOLIN(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Solar insolation
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SWCF(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Shortwave cloud forcing
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - T(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- K
- long_name :
- Temperature
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TAUGWX(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- N/m2
- long_name :
- Zonal gravity wave surface stress
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TAUGWY(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- N/m2
- long_name :
- Meridional gravity wave surface stress
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TAUX(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- N/m2
- long_name :
- Zonal surface stress
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TAUY(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- N/m2
- long_name :
- Meridional surface stress
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TGCLDCWP(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- gram/m2
- long_name :
- Total grid-box cloud water path (liquid and ice)
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TGCLDIWP(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- gram/m2
- long_name :
- Total grid-box cloud ice water path
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TGCLDLWP(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- Sampling_Sequence :
- rad_lwsw
- units :
- gram/m2
- long_name :
- Total grid-box cloud liquid water path
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TH(time, ilev, lat, lon)float32dask.array<chunksize=(1, 27, 96, 144), meta=np.ndarray>
- mdims :
- 2
- units :
- K
- long_name :
- Potential Temperature
- cell_methods :
- time: mean
Array Chunk Bytes 17.09 MiB 1.42 MiB Shape (12, 27, 96, 144) (1, 27, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - THzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- K
- long_name :
- Zonal-Mean potential temp - defined on ilev
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TMQ(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- kg/m2
- long_name :
- Total (vertically integrated) precipitable water
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TREFHT(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- K
- long_name :
- Reference height temperature
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TS(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- K
- long_name :
- Surface temperature (radiative)
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TSMN(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- K
- long_name :
- Minimum surface temperature over output period
- cell_methods :
- time: minimum
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TSMX(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- K
- long_name :
- Maximum surface temperature over output period
- cell_methods :
- time: maximum
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - U(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m/s
- long_name :
- Zonal wind
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - U10(time, lat, lon)float32dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
- units :
- m/s
- long_name :
- 10m wind speed
- cell_methods :
- time: mean
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - UTGWORO(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m/s2
- long_name :
- U tendency - orographic gravity wave drag
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - UU(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m2/s2
- long_name :
- Zonal velocity squared
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - UVzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- M2/S2
- long_name :
- Meridional Flux of Zonal Momentum: 3D zon. mean
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - UWzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- M2/S2
- long_name :
- Vertical Flux of Zonal Momentum: 3D zon. mean
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - Uzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- M/S
- long_name :
- Zonal-Mean zonal wind - defined on ilev
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - V(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m/s
- long_name :
- Meridional wind
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VD01(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- kg/kg/s
- long_name :
- Vertical diffusion of Q
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VQ(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m/skg/kg
- long_name :
- Meridional water transport
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VT(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- K m/s
- long_name :
- Meridional heat transport
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VTHzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- MK/S
- long_name :
- Meridional Heat Flux: 3D zon. mean
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VU(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m2/s2
- long_name :
- Meridional flux of zonal momentum
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VV(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m2/s2
- long_name :
- Meridional velocity squared
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - Vzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- M/S
- long_name :
- Zonal-Mean meridional wind - defined on ilev
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - WTHzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- MK/S
- long_name :
- Vertical Heat Flux: 3D zon. mean
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - Wzm(time, ilev, lat, zlon)float32dask.array<chunksize=(1, 27, 96, 1), meta=np.ndarray>
- mdims :
- 2
- units :
- M/S
- long_name :
- Zonal-Mean vertical wind - defined on ilev
- cell_methods :
- zlon: mean time: mean
Array Chunk Bytes 121.50 kiB 10.12 kiB Shape (12, 27, 96, 1) (1, 27, 96, 1) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - Z3(time, lev, lat, lon)float32dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
- mdims :
- 1
- units :
- m
- long_name :
- Geopotential Height (above sea level)
- cell_methods :
- time: mean
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray
- latPandasIndex
PandasIndex(Index([ -90.0, -88.10526315789474, -86.21052631578948, -84.3157894736842, -82.42105263157895, -80.52631578947368, -78.63157894736842, -76.73684210526316, -74.84210526315789, -72.94736842105263, -71.05263157894737, -69.15789473684211, -67.26315789473685, -65.36842105263158, -63.473684210526315, -61.578947368421055, -59.684210526315795, -57.78947368421053, -55.89473684210527, -54.0, -52.10526315789474, -50.21052631578947, -48.31578947368421, -46.42105263157895, -44.526315789473685, -42.631578947368425, -40.73684210526316, -38.8421052631579, -36.94736842105264, -35.05263157894737, -33.15789473684211, -31.263157894736842, -29.368421052631582, -27.473684210526322, -25.578947368421055, -23.684210526315795, -21.789473684210535, -19.89473684210526, -18.0, -16.10526315789474, -14.21052631578948, -12.31578947368422, -10.421052631578945, -8.526315789473685, -6.631578947368425, -4.736842105263165, -2.8421052631579045, -0.9473684210526301, 0.9473684210526301, 2.8421052631578902, 4.73684210526315, 6.631578947368411, 8.526315789473685, 10.421052631578945, 12.315789473684205, 14.210526315789465, 16.105263157894726, 18.0, 19.89473684210526, 21.78947368421052, 23.68421052631578, 25.57894736842104, 27.473684210526315, 29.368421052631575, 31.263157894736835, 33.157894736842096, 35.052631578947356, 36.94736842105263, 38.84210526315789, 40.73684210526315, 42.63157894736841, 44.52631578947367, 46.42105263157893, 48.31578947368419, 50.21052631578948, 52.10526315789474, 54.0, 55.89473684210526, 57.78947368421052, 59.68421052631578, 61.57894736842104, 63.4736842105263, 65.36842105263156, 67.26315789473682, 69.15789473684211, 71.05263157894737, 72.94736842105263, 74.84210526315789, 76.73684210526315, 78.63157894736841, 80.52631578947367, 82.42105263157893, 84.31578947368419, 86.21052631578945, 88.10526315789474, 90.0], dtype='float64', name='lat')) - zlonPandasIndex
PandasIndex(Index([0.0], dtype='float64', name='zlon'))
- lonPandasIndex
PandasIndex(Index([ 0.0, 2.5, 5.0, 7.5, 10.0, 12.5, 15.0, 17.5, 20.0, 22.5, ... 335.0, 337.5, 340.0, 342.5, 345.0, 347.5, 350.0, 352.5, 355.0, 357.5], dtype='float64', name='lon', length=144)) - levPandasIndex
PandasIndex(Index([3.5446380000000097, 7.3888135000000075, 13.967214000000006, 23.944625, 37.23029000000011, 53.1146050000002, 70.05915000000029, 85.43911500000031, 100.51469500000029, 118.25033500000026, 139.11539500000046, 163.66207000000043, 192.53993500000033, 226.51326500000036, 266.4811550000001, 313.5012650000006, 368.81798000000157, 433.8952250000011, 510.45525500000167, 600.5242000000027, 696.7962900000033, 787.7020600000026, 867.1607600000013, 929.6488750000024, 970.5548300000014, 992.5560999999998], dtype='float64', name='lev')) - ilevPandasIndex
PandasIndex(Index([ 2.194067000000008, 4.895209000000012, 9.882418000000005, 18.052010000000006, 29.837239999999987, 44.62334000000023, 61.60587000000017, 78.5124300000004, 92.36580000000022, 108.66359000000037, 127.83708000000016, 150.3937100000008, 176.9304300000001, 208.14944000000057, 244.87709000000012, 288.08522000000016, 338.9173100000011, 398.71865000000196, 469.0718000000003, 551.8387100000031, 649.2096900000024, 744.3828900000042, 831.0212300000011, 903.3002900000016, 955.9974600000032, 985.1121999999997, 1000.0], dtype='float64', name='ilev')) - timePandasIndex
PandasIndex(CFTimeIndex([0001-01-17 00:00:00, 0001-02-14 00:00:00, 0001-03-17 00:00:00, 0001-04-16 00:00:00, 0001-05-17 00:00:00, 0001-06-16 00:00:00, 0001-07-17 00:00:00, 0001-08-17 00:00:00, 0001-09-16 00:00:00, 0001-10-17 00:00:00, 0001-11-16 00:00:00, 0001-12-17 00:00:00], dtype='object', length=12, calendar='noleap', freq=None))
- Conventions :
- CF-1.0
- source :
- CAM
- case :
- b.e21.B1850.f19_g17.piControl.001
- logname :
- jiangzhu
- host :
- derecho2
- initial_file :
- /glade/campaign/cesm/cesmdata/inputdata/atm/cam/inic/fv/cami_0000-01-01_1.9x2.5_L26_c070408.nc
- topography_file :
- /glade/campaign/cesm/cesmdata/inputdata/atm/cam/topo/fv_1.9x2.5_nc3000_Nsw084_Nrs016_Co120_Fi001_ZR_GRNL_031819.nc
- model_doi_url :
- https://doi.org/10.5065/D67H1H0V
- time_period_freq :
- month_1
Compute the zonal mean of solar insolation and make plots#
Examine a variable including its dimension, long name, and units
Use the
.iselmethod to select a signle month or multiple monthsUse the Xarray plotting functionality to make a simple plot
Use the hvplot to make an interactive plot
Use
.mean('lon')to get the zonal mean
ds_PI.SOLIN
<xarray.DataArray 'SOLIN' (time: 12, lat: 96, lon: 144)> Size: 664kB
dask.array<concatenate, shape=(12, 96, 144), dtype=float32, chunksize=(1, 96, 144), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float64 768B -90.0 -88.11 -86.21 -84.32 ... 86.21 88.11 90.0
* lon (lon) float64 1kB 0.0 2.5 5.0 7.5 10.0 ... 350.0 352.5 355.0 357.5
* time (time) object 96B 0001-01-17 00:00:00 ... 0001-12-17 00:00:00
Attributes:
Sampling_Sequence: rad_lwsw
units: W/m2
long_name: Solar insolation
cell_methods: time: mean- time: 12
- lat: 96
- lon: 144
- dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - lat(lat)float64-90.0 -88.11 -86.21 ... 88.11 90.0
- long_name :
- latitude
- units :
- degrees_north
array([-90. , -88.105263, -86.210526, -84.315789, -82.421053, -80.526316, -78.631579, -76.736842, -74.842105, -72.947368, -71.052632, -69.157895, -67.263158, -65.368421, -63.473684, -61.578947, -59.684211, -57.789474, -55.894737, -54. , -52.105263, -50.210526, -48.315789, -46.421053, -44.526316, -42.631579, -40.736842, -38.842105, -36.947368, -35.052632, -33.157895, -31.263158, -29.368421, -27.473684, -25.578947, -23.684211, -21.789474, -19.894737, -18. , -16.105263, -14.210526, -12.315789, -10.421053, -8.526316, -6.631579, -4.736842, -2.842105, -0.947368, 0.947368, 2.842105, 4.736842, 6.631579, 8.526316, 10.421053, 12.315789, 14.210526, 16.105263, 18. , 19.894737, 21.789474, 23.684211, 25.578947, 27.473684, 29.368421, 31.263158, 33.157895, 35.052632, 36.947368, 38.842105, 40.736842, 42.631579, 44.526316, 46.421053, 48.315789, 50.210526, 52.105263, 54. , 55.894737, 57.789474, 59.684211, 61.578947, 63.473684, 65.368421, 67.263158, 69.157895, 71.052632, 72.947368, 74.842105, 76.736842, 78.631579, 80.526316, 82.421053, 84.315789, 86.210526, 88.105263, 90. ]) - lon(lon)float640.0 2.5 5.0 ... 352.5 355.0 357.5
- long_name :
- longitude
- units :
- degrees_east
array([ 0. , 2.5, 5. , 7.5, 10. , 12.5, 15. , 17.5, 20. , 22.5, 25. , 27.5, 30. , 32.5, 35. , 37.5, 40. , 42.5, 45. , 47.5, 50. , 52.5, 55. , 57.5, 60. , 62.5, 65. , 67.5, 70. , 72.5, 75. , 77.5, 80. , 82.5, 85. , 87.5, 90. , 92.5, 95. , 97.5, 100. , 102.5, 105. , 107.5, 110. , 112.5, 115. , 117.5, 120. , 122.5, 125. , 127.5, 130. , 132.5, 135. , 137.5, 140. , 142.5, 145. , 147.5, 150. , 152.5, 155. , 157.5, 160. , 162.5, 165. , 167.5, 170. , 172.5, 175. , 177.5, 180. , 182.5, 185. , 187.5, 190. , 192.5, 195. , 197.5, 200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. , 332.5, 335. , 337.5, 340. , 342.5, 345. , 347.5, 350. , 352.5, 355. , 357.5]) - time(time)object0001-01-17 00:00:00 ... 0001-12-...
array([cftime.DatetimeNoLeap(1, 1, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 2, 14, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 3, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 4, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 5, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 6, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 7, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 8, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 9, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 10, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 12, 17, 0, 0, 0, 0, has_year_zero=True)], dtype=object)
- latPandasIndex
PandasIndex(Index([ -90.0, -88.10526315789474, -86.21052631578948, -84.3157894736842, -82.42105263157895, -80.52631578947368, -78.63157894736842, -76.73684210526316, -74.84210526315789, -72.94736842105263, -71.05263157894737, -69.15789473684211, -67.26315789473685, -65.36842105263158, -63.473684210526315, -61.578947368421055, -59.684210526315795, -57.78947368421053, -55.89473684210527, -54.0, -52.10526315789474, -50.21052631578947, -48.31578947368421, -46.42105263157895, -44.526315789473685, -42.631578947368425, -40.73684210526316, -38.8421052631579, -36.94736842105264, -35.05263157894737, -33.15789473684211, -31.263157894736842, -29.368421052631582, -27.473684210526322, -25.578947368421055, -23.684210526315795, -21.789473684210535, -19.89473684210526, -18.0, -16.10526315789474, -14.21052631578948, -12.31578947368422, -10.421052631578945, -8.526315789473685, -6.631578947368425, -4.736842105263165, -2.8421052631579045, -0.9473684210526301, 0.9473684210526301, 2.8421052631578902, 4.73684210526315, 6.631578947368411, 8.526315789473685, 10.421052631578945, 12.315789473684205, 14.210526315789465, 16.105263157894726, 18.0, 19.89473684210526, 21.78947368421052, 23.68421052631578, 25.57894736842104, 27.473684210526315, 29.368421052631575, 31.263157894736835, 33.157894736842096, 35.052631578947356, 36.94736842105263, 38.84210526315789, 40.73684210526315, 42.63157894736841, 44.52631578947367, 46.42105263157893, 48.31578947368419, 50.21052631578948, 52.10526315789474, 54.0, 55.89473684210526, 57.78947368421052, 59.68421052631578, 61.57894736842104, 63.4736842105263, 65.36842105263156, 67.26315789473682, 69.15789473684211, 71.05263157894737, 72.94736842105263, 74.84210526315789, 76.73684210526315, 78.63157894736841, 80.52631578947367, 82.42105263157893, 84.31578947368419, 86.21052631578945, 88.10526315789474, 90.0], dtype='float64', name='lat')) - lonPandasIndex
PandasIndex(Index([ 0.0, 2.5, 5.0, 7.5, 10.0, 12.5, 15.0, 17.5, 20.0, 22.5, ... 335.0, 337.5, 340.0, 342.5, 345.0, 347.5, 350.0, 352.5, 355.0, 357.5], dtype='float64', name='lon', length=144)) - timePandasIndex
PandasIndex(CFTimeIndex([0001-01-17 00:00:00, 0001-02-14 00:00:00, 0001-03-17 00:00:00, 0001-04-16 00:00:00, 0001-05-17 00:00:00, 0001-06-16 00:00:00, 0001-07-17 00:00:00, 0001-08-17 00:00:00, 0001-09-16 00:00:00, 0001-10-17 00:00:00, 0001-11-16 00:00:00, 0001-12-17 00:00:00], dtype='object', length=12, calendar='noleap', freq=None))
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Solar insolation
- cell_methods :
- time: mean
ds_PI.SOLIN.isel(time=5).plot(size=2)
/glade/u/apps/opt/conda/envs/npl-2025a/lib/python3.12/site-packages/dask/config.py:787: FutureWarning: Dask configuration key 'allowed-failures' has been deprecated; please use 'distributed.scheduler.allowed-failures' instead
warnings.warn(
<matplotlib.collections.QuadMesh at 0x1489712bfbc0>
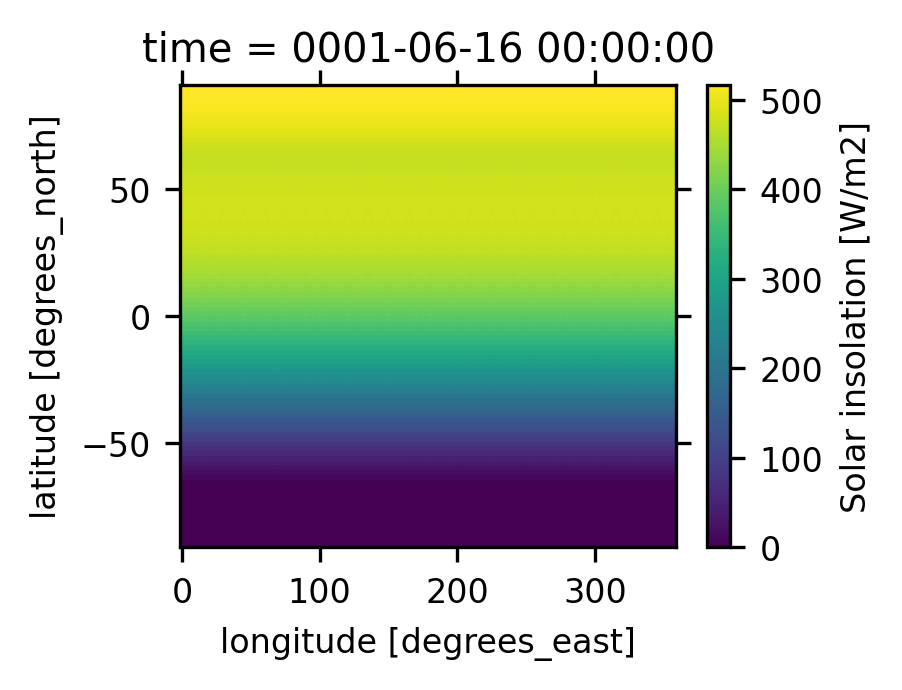
# Easier to find value for SNARL (37.6N, -118.8E)
ds_PI.SOLIN.isel(time=5).hvplot(coastline=True, cmap='viridis')
ds_PI.SOLIN
<xarray.DataArray 'SOLIN' (time: 12, lat: 96, lon: 144)> Size: 664kB
dask.array<concatenate, shape=(12, 96, 144), dtype=float32, chunksize=(1, 96, 144), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float64 768B -90.0 -88.11 -86.21 -84.32 ... 86.21 88.11 90.0
* lon (lon) float64 1kB 0.0 2.5 5.0 7.5 10.0 ... 350.0 352.5 355.0 357.5
* time (time) object 96B 0001-01-17 00:00:00 ... 0001-12-17 00:00:00
Attributes:
Sampling_Sequence: rad_lwsw
units: W/m2
long_name: Solar insolation
cell_methods: time: mean- time: 12
- lat: 96
- lon: 144
- dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - lat(lat)float64-90.0 -88.11 -86.21 ... 88.11 90.0
- long_name :
- latitude
- units :
- degrees_north
array([-90. , -88.105263, -86.210526, -84.315789, -82.421053, -80.526316, -78.631579, -76.736842, -74.842105, -72.947368, -71.052632, -69.157895, -67.263158, -65.368421, -63.473684, -61.578947, -59.684211, -57.789474, -55.894737, -54. , -52.105263, -50.210526, -48.315789, -46.421053, -44.526316, -42.631579, -40.736842, -38.842105, -36.947368, -35.052632, -33.157895, -31.263158, -29.368421, -27.473684, -25.578947, -23.684211, -21.789474, -19.894737, -18. , -16.105263, -14.210526, -12.315789, -10.421053, -8.526316, -6.631579, -4.736842, -2.842105, -0.947368, 0.947368, 2.842105, 4.736842, 6.631579, 8.526316, 10.421053, 12.315789, 14.210526, 16.105263, 18. , 19.894737, 21.789474, 23.684211, 25.578947, 27.473684, 29.368421, 31.263158, 33.157895, 35.052632, 36.947368, 38.842105, 40.736842, 42.631579, 44.526316, 46.421053, 48.315789, 50.210526, 52.105263, 54. , 55.894737, 57.789474, 59.684211, 61.578947, 63.473684, 65.368421, 67.263158, 69.157895, 71.052632, 72.947368, 74.842105, 76.736842, 78.631579, 80.526316, 82.421053, 84.315789, 86.210526, 88.105263, 90. ]) - lon(lon)float640.0 2.5 5.0 ... 352.5 355.0 357.5
- long_name :
- longitude
- units :
- degrees_east
array([ 0. , 2.5, 5. , 7.5, 10. , 12.5, 15. , 17.5, 20. , 22.5, 25. , 27.5, 30. , 32.5, 35. , 37.5, 40. , 42.5, 45. , 47.5, 50. , 52.5, 55. , 57.5, 60. , 62.5, 65. , 67.5, 70. , 72.5, 75. , 77.5, 80. , 82.5, 85. , 87.5, 90. , 92.5, 95. , 97.5, 100. , 102.5, 105. , 107.5, 110. , 112.5, 115. , 117.5, 120. , 122.5, 125. , 127.5, 130. , 132.5, 135. , 137.5, 140. , 142.5, 145. , 147.5, 150. , 152.5, 155. , 157.5, 160. , 162.5, 165. , 167.5, 170. , 172.5, 175. , 177.5, 180. , 182.5, 185. , 187.5, 190. , 192.5, 195. , 197.5, 200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. , 332.5, 335. , 337.5, 340. , 342.5, 345. , 347.5, 350. , 352.5, 355. , 357.5]) - time(time)object0001-01-17 00:00:00 ... 0001-12-...
array([cftime.DatetimeNoLeap(1, 1, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 2, 14, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 3, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 4, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 5, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 6, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 7, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 8, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 9, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 10, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 12, 17, 0, 0, 0, 0, has_year_zero=True)], dtype=object)
- latPandasIndex
PandasIndex(Index([ -90.0, -88.10526315789474, -86.21052631578948, -84.3157894736842, -82.42105263157895, -80.52631578947368, -78.63157894736842, -76.73684210526316, -74.84210526315789, -72.94736842105263, -71.05263157894737, -69.15789473684211, -67.26315789473685, -65.36842105263158, -63.473684210526315, -61.578947368421055, -59.684210526315795, -57.78947368421053, -55.89473684210527, -54.0, -52.10526315789474, -50.21052631578947, -48.31578947368421, -46.42105263157895, -44.526315789473685, -42.631578947368425, -40.73684210526316, -38.8421052631579, -36.94736842105264, -35.05263157894737, -33.15789473684211, -31.263157894736842, -29.368421052631582, -27.473684210526322, -25.578947368421055, -23.684210526315795, -21.789473684210535, -19.89473684210526, -18.0, -16.10526315789474, -14.21052631578948, -12.31578947368422, -10.421052631578945, -8.526315789473685, -6.631578947368425, -4.736842105263165, -2.8421052631579045, -0.9473684210526301, 0.9473684210526301, 2.8421052631578902, 4.73684210526315, 6.631578947368411, 8.526315789473685, 10.421052631578945, 12.315789473684205, 14.210526315789465, 16.105263157894726, 18.0, 19.89473684210526, 21.78947368421052, 23.68421052631578, 25.57894736842104, 27.473684210526315, 29.368421052631575, 31.263157894736835, 33.157894736842096, 35.052631578947356, 36.94736842105263, 38.84210526315789, 40.73684210526315, 42.63157894736841, 44.52631578947367, 46.42105263157893, 48.31578947368419, 50.21052631578948, 52.10526315789474, 54.0, 55.89473684210526, 57.78947368421052, 59.68421052631578, 61.57894736842104, 63.4736842105263, 65.36842105263156, 67.26315789473682, 69.15789473684211, 71.05263157894737, 72.94736842105263, 74.84210526315789, 76.73684210526315, 78.63157894736841, 80.52631578947367, 82.42105263157893, 84.31578947368419, 86.21052631578945, 88.10526315789474, 90.0], dtype='float64', name='lat')) - lonPandasIndex
PandasIndex(Index([ 0.0, 2.5, 5.0, 7.5, 10.0, 12.5, 15.0, 17.5, 20.0, 22.5, ... 335.0, 337.5, 340.0, 342.5, 345.0, 347.5, 350.0, 352.5, 355.0, 357.5], dtype='float64', name='lon', length=144)) - timePandasIndex
PandasIndex(CFTimeIndex([0001-01-17 00:00:00, 0001-02-14 00:00:00, 0001-03-17 00:00:00, 0001-04-16 00:00:00, 0001-05-17 00:00:00, 0001-06-16 00:00:00, 0001-07-17 00:00:00, 0001-08-17 00:00:00, 0001-09-16 00:00:00, 0001-10-17 00:00:00, 0001-11-16 00:00:00, 0001-12-17 00:00:00], dtype='object', length=12, calendar='noleap', freq=None))
- Sampling_Sequence :
- rad_lwsw
- units :
- W/m2
- long_name :
- Solar insolation
- cell_methods :
- time: mean
# Compute zonal mean and make plot of piControl
ds_PI.SOLIN.mean('lon').plot.contourf(x='time', y='lat', figsize=(4, 2))
plt.xlabel('time (year-month)')
Text(0.5, 0, 'time (year-month)')
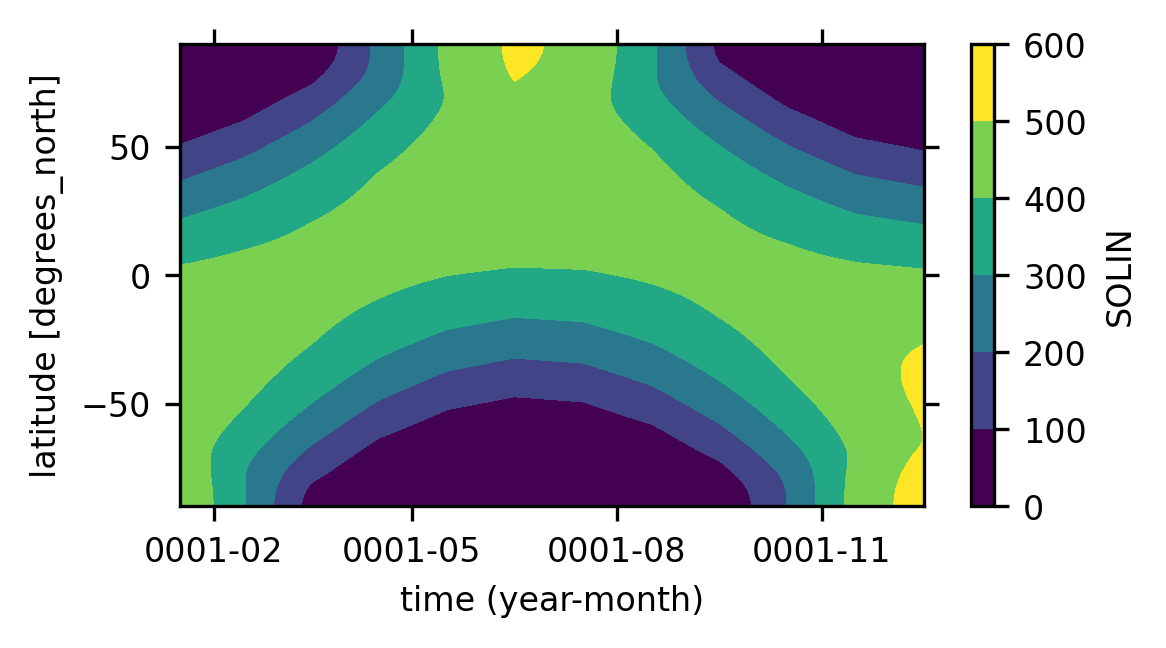
# Add your code to compute zonal mean and make plot of midHolocene
# Add your code to compute and plot the difference: midHolocen - piControl zonal mean
Click here for the solution
Copy and paste the code into the above cell
ds_MH.SOLIN.mean('lon').plot.contourf(x='time', y='lat', figsize=(4, 2))
(ds_MH.SOLIN - ds_PI.SOLIN).mean('lon').plot.contourf(
x='time', y='lat', figsize=(4, 2), levels=np.linspace(-30, 30, 21))
Small group discussion#
Which orbital parameters are different at the middle Holocene (6ka BP)?
How does the orbital parameter impact the top-of-atmosphere shortwave radiation (solar insolation)
Do the results look correct? You can compare your results with Figure 3b of Otto-Bliesner et al., (2017)
Analysis 2: Does the Earth receive more radiation during the midHolocene?#
We compute the global annual mean solar insolation to answer this question.
Importantly, we need to weight grid cells by their area (equivalent to cosine of latitude), using the
.weightedmethod
Note that lat is in degree
ds_PI.lat
<xarray.DataArray 'lat' (lat: 96)> Size: 768B
array([-90. , -88.105263, -86.210526, -84.315789, -82.421053, -80.526316,
-78.631579, -76.736842, -74.842105, -72.947368, -71.052632, -69.157895,
-67.263158, -65.368421, -63.473684, -61.578947, -59.684211, -57.789474,
-55.894737, -54. , -52.105263, -50.210526, -48.315789, -46.421053,
-44.526316, -42.631579, -40.736842, -38.842105, -36.947368, -35.052632,
-33.157895, -31.263158, -29.368421, -27.473684, -25.578947, -23.684211,
-21.789474, -19.894737, -18. , -16.105263, -14.210526, -12.315789,
-10.421053, -8.526316, -6.631579, -4.736842, -2.842105, -0.947368,
0.947368, 2.842105, 4.736842, 6.631579, 8.526316, 10.421053,
12.315789, 14.210526, 16.105263, 18. , 19.894737, 21.789474,
23.684211, 25.578947, 27.473684, 29.368421, 31.263158, 33.157895,
35.052632, 36.947368, 38.842105, 40.736842, 42.631579, 44.526316,
46.421053, 48.315789, 50.210526, 52.105263, 54. , 55.894737,
57.789474, 59.684211, 61.578947, 63.473684, 65.368421, 67.263158,
69.157895, 71.052632, 72.947368, 74.842105, 76.736842, 78.631579,
80.526316, 82.421053, 84.315789, 86.210526, 88.105263, 90. ])
Coordinates:
* lat (lat) float64 768B -90.0 -88.11 -86.21 -84.32 ... 86.21 88.11 90.0
Attributes:
long_name: latitude
units: degrees_north- lat: 96
- -90.0 -88.11 -86.21 -84.32 -82.42 ... 82.42 84.32 86.21 88.11 90.0
array([-90. , -88.105263, -86.210526, -84.315789, -82.421053, -80.526316, -78.631579, -76.736842, -74.842105, -72.947368, -71.052632, -69.157895, -67.263158, -65.368421, -63.473684, -61.578947, -59.684211, -57.789474, -55.894737, -54. , -52.105263, -50.210526, -48.315789, -46.421053, -44.526316, -42.631579, -40.736842, -38.842105, -36.947368, -35.052632, -33.157895, -31.263158, -29.368421, -27.473684, -25.578947, -23.684211, -21.789474, -19.894737, -18. , -16.105263, -14.210526, -12.315789, -10.421053, -8.526316, -6.631579, -4.736842, -2.842105, -0.947368, 0.947368, 2.842105, 4.736842, 6.631579, 8.526316, 10.421053, 12.315789, 14.210526, 16.105263, 18. , 19.894737, 21.789474, 23.684211, 25.578947, 27.473684, 29.368421, 31.263158, 33.157895, 35.052632, 36.947368, 38.842105, 40.736842, 42.631579, 44.526316, 46.421053, 48.315789, 50.210526, 52.105263, 54. , 55.894737, 57.789474, 59.684211, 61.578947, 63.473684, 65.368421, 67.263158, 69.157895, 71.052632, 72.947368, 74.842105, 76.736842, 78.631579, 80.526316, 82.421053, 84.315789, 86.210526, 88.105263, 90. ]) - lat(lat)float64-90.0 -88.11 -86.21 ... 88.11 90.0
- long_name :
- latitude
- units :
- degrees_north
array([-90. , -88.105263, -86.210526, -84.315789, -82.421053, -80.526316, -78.631579, -76.736842, -74.842105, -72.947368, -71.052632, -69.157895, -67.263158, -65.368421, -63.473684, -61.578947, -59.684211, -57.789474, -55.894737, -54. , -52.105263, -50.210526, -48.315789, -46.421053, -44.526316, -42.631579, -40.736842, -38.842105, -36.947368, -35.052632, -33.157895, -31.263158, -29.368421, -27.473684, -25.578947, -23.684211, -21.789474, -19.894737, -18. , -16.105263, -14.210526, -12.315789, -10.421053, -8.526316, -6.631579, -4.736842, -2.842105, -0.947368, 0.947368, 2.842105, 4.736842, 6.631579, 8.526316, 10.421053, 12.315789, 14.210526, 16.105263, 18. , 19.894737, 21.789474, 23.684211, 25.578947, 27.473684, 29.368421, 31.263158, 33.157895, 35.052632, 36.947368, 38.842105, 40.736842, 42.631579, 44.526316, 46.421053, 48.315789, 50.210526, 52.105263, 54. , 55.894737, 57.789474, 59.684211, 61.578947, 63.473684, 65.368421, 67.263158, 69.157895, 71.052632, 72.947368, 74.842105, 76.736842, 78.631579, 80.526316, 82.421053, 84.315789, 86.210526, 88.105263, 90. ])
- latPandasIndex
PandasIndex(Index([ -90.0, -88.10526315789474, -86.21052631578948, -84.3157894736842, -82.42105263157895, -80.52631578947368, -78.63157894736842, -76.73684210526316, -74.84210526315789, -72.94736842105263, -71.05263157894737, -69.15789473684211, -67.26315789473685, -65.36842105263158, -63.473684210526315, -61.578947368421055, -59.684210526315795, -57.78947368421053, -55.89473684210527, -54.0, -52.10526315789474, -50.21052631578947, -48.31578947368421, -46.42105263157895, -44.526315789473685, -42.631578947368425, -40.73684210526316, -38.8421052631579, -36.94736842105264, -35.05263157894737, -33.15789473684211, -31.263157894736842, -29.368421052631582, -27.473684210526322, -25.578947368421055, -23.684210526315795, -21.789473684210535, -19.89473684210526, -18.0, -16.10526315789474, -14.21052631578948, -12.31578947368422, -10.421052631578945, -8.526315789473685, -6.631578947368425, -4.736842105263165, -2.8421052631579045, -0.9473684210526301, 0.9473684210526301, 2.8421052631578902, 4.73684210526315, 6.631578947368411, 8.526315789473685, 10.421052631578945, 12.315789473684205, 14.210526315789465, 16.105263157894726, 18.0, 19.89473684210526, 21.78947368421052, 23.68421052631578, 25.57894736842104, 27.473684210526315, 29.368421052631575, 31.263157894736835, 33.157894736842096, 35.052631578947356, 36.94736842105263, 38.84210526315789, 40.73684210526315, 42.63157894736841, 44.52631578947367, 46.42105263157893, 48.31578947368419, 50.21052631578948, 52.10526315789474, 54.0, 55.89473684210526, 57.78947368421052, 59.68421052631578, 61.57894736842104, 63.4736842105263, 65.36842105263156, 67.26315789473682, 69.15789473684211, 71.05263157894737, 72.94736842105263, 74.84210526315789, 76.73684210526315, 78.63157894736841, 80.52631578947367, 82.42105263157893, 84.31578947368419, 86.21052631578945, 88.10526315789474, 90.0], dtype='float64', name='lat'))
- long_name :
- latitude
- units :
- degrees_north
Example calculation for the piControl#
coslat = np.cos(np.deg2rad(ds_PI.lat))
SOLIN_PI = ds_PI.SOLIN.weighted(coslat).mean(('lat', 'lon', 'time'))
print("The global annual mean insolation of PI: ", SOLIN_PI.values, "W/m2")
The global annual mean insolation of PI: 340.3271967819253 W/m2
If we don’t apply the area weights, we will get a wrong answer#
SOLIN_PI_unweighted = ds_PI.SOLIN.mean(('lat', 'lon', 'time'))
print("Only if we forgot to properly weight the values by area (cosine(latitude)):",
SOLIN_PI_unweighted.values, "W/m2")
Only if we forgot to properly weight the values by area (cosine(latitude)): 297.03168 W/m2
Add your own calculation for the mid-Holocene#
Click here for the solution
Copy and paste the code into the above cell
coslat = np.cos(np.deg2rad(ds_MH.lat))
SOLIN_MH = ds_MH.SOLIN.weighted(coslat).mean(('lat', 'lon', 'time'))
print("The global annual mean insolation of MH: ", SOLIN_MH.values, "W/m2")
print("Difference, MH - PI: ", SOLIN_MH.values-SOLIN_PI.values, "W/m2")
Small group discussion#
Should the mid-Holocene be warmer or colder than the preindustrial?
Geological records suggest that MH may be warmer than the present day
Climate models in general suggest a colder mid-Holocene
Further reading: Osman et al. (2021)
Ask Jess and Jiang about the Holocene Temperature Conundrum
Analysis 3: how does orbital forcing impact the ITCZ and monsoon precipitation during the midHolocene?#
In CESM, total precipitation is computed as
prec = PRECC + PRECL(sum of convective and large-scale precipitation; recall that climate models have to parameterize convection!)We convert the units from m/s into mm/day
We use
.isel(time=slice(5, 8))to select the June, July, and Agugust values (recall that Python uses 0-based ordering)Let’s use Cartopy and Matplotlib with a Robinson projection (instead of using the simple Xarray.plot())
We use
add_cyclic_pointto get rid of the “white strip” in the plot
ds_PI.PRECC
<xarray.DataArray 'PRECC' (time: 12, lat: 96, lon: 144)> Size: 664kB
dask.array<concatenate, shape=(12, 96, 144), dtype=float32, chunksize=(1, 96, 144), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float64 768B -90.0 -88.11 -86.21 -84.32 ... 86.21 88.11 90.0
* lon (lon) float64 1kB 0.0 2.5 5.0 7.5 10.0 ... 350.0 352.5 355.0 357.5
* time (time) object 96B 0001-01-17 00:00:00 ... 0001-12-17 00:00:00
Attributes:
units: m/s
long_name: Convective precipitation rate (liq + ice)
cell_methods: time: mean- time: 12
- lat: 96
- lon: 144
- dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - lat(lat)float64-90.0 -88.11 -86.21 ... 88.11 90.0
- long_name :
- latitude
- units :
- degrees_north
array([-90. , -88.105263, -86.210526, -84.315789, -82.421053, -80.526316, -78.631579, -76.736842, -74.842105, -72.947368, -71.052632, -69.157895, -67.263158, -65.368421, -63.473684, -61.578947, -59.684211, -57.789474, -55.894737, -54. , -52.105263, -50.210526, -48.315789, -46.421053, -44.526316, -42.631579, -40.736842, -38.842105, -36.947368, -35.052632, -33.157895, -31.263158, -29.368421, -27.473684, -25.578947, -23.684211, -21.789474, -19.894737, -18. , -16.105263, -14.210526, -12.315789, -10.421053, -8.526316, -6.631579, -4.736842, -2.842105, -0.947368, 0.947368, 2.842105, 4.736842, 6.631579, 8.526316, 10.421053, 12.315789, 14.210526, 16.105263, 18. , 19.894737, 21.789474, 23.684211, 25.578947, 27.473684, 29.368421, 31.263158, 33.157895, 35.052632, 36.947368, 38.842105, 40.736842, 42.631579, 44.526316, 46.421053, 48.315789, 50.210526, 52.105263, 54. , 55.894737, 57.789474, 59.684211, 61.578947, 63.473684, 65.368421, 67.263158, 69.157895, 71.052632, 72.947368, 74.842105, 76.736842, 78.631579, 80.526316, 82.421053, 84.315789, 86.210526, 88.105263, 90. ]) - lon(lon)float640.0 2.5 5.0 ... 352.5 355.0 357.5
- long_name :
- longitude
- units :
- degrees_east
array([ 0. , 2.5, 5. , 7.5, 10. , 12.5, 15. , 17.5, 20. , 22.5, 25. , 27.5, 30. , 32.5, 35. , 37.5, 40. , 42.5, 45. , 47.5, 50. , 52.5, 55. , 57.5, 60. , 62.5, 65. , 67.5, 70. , 72.5, 75. , 77.5, 80. , 82.5, 85. , 87.5, 90. , 92.5, 95. , 97.5, 100. , 102.5, 105. , 107.5, 110. , 112.5, 115. , 117.5, 120. , 122.5, 125. , 127.5, 130. , 132.5, 135. , 137.5, 140. , 142.5, 145. , 147.5, 150. , 152.5, 155. , 157.5, 160. , 162.5, 165. , 167.5, 170. , 172.5, 175. , 177.5, 180. , 182.5, 185. , 187.5, 190. , 192.5, 195. , 197.5, 200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. , 332.5, 335. , 337.5, 340. , 342.5, 345. , 347.5, 350. , 352.5, 355. , 357.5]) - time(time)object0001-01-17 00:00:00 ... 0001-12-...
array([cftime.DatetimeNoLeap(1, 1, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 2, 14, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 3, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 4, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 5, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 6, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 7, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 8, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 9, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 10, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 12, 17, 0, 0, 0, 0, has_year_zero=True)], dtype=object)
- latPandasIndex
PandasIndex(Index([ -90.0, -88.10526315789474, -86.21052631578948, -84.3157894736842, -82.42105263157895, -80.52631578947368, -78.63157894736842, -76.73684210526316, -74.84210526315789, -72.94736842105263, -71.05263157894737, -69.15789473684211, -67.26315789473685, -65.36842105263158, -63.473684210526315, -61.578947368421055, -59.684210526315795, -57.78947368421053, -55.89473684210527, -54.0, -52.10526315789474, -50.21052631578947, -48.31578947368421, -46.42105263157895, -44.526315789473685, -42.631578947368425, -40.73684210526316, -38.8421052631579, -36.94736842105264, -35.05263157894737, -33.15789473684211, -31.263157894736842, -29.368421052631582, -27.473684210526322, -25.578947368421055, -23.684210526315795, -21.789473684210535, -19.89473684210526, -18.0, -16.10526315789474, -14.21052631578948, -12.31578947368422, -10.421052631578945, -8.526315789473685, -6.631578947368425, -4.736842105263165, -2.8421052631579045, -0.9473684210526301, 0.9473684210526301, 2.8421052631578902, 4.73684210526315, 6.631578947368411, 8.526315789473685, 10.421052631578945, 12.315789473684205, 14.210526315789465, 16.105263157894726, 18.0, 19.89473684210526, 21.78947368421052, 23.68421052631578, 25.57894736842104, 27.473684210526315, 29.368421052631575, 31.263157894736835, 33.157894736842096, 35.052631578947356, 36.94736842105263, 38.84210526315789, 40.73684210526315, 42.63157894736841, 44.52631578947367, 46.42105263157893, 48.31578947368419, 50.21052631578948, 52.10526315789474, 54.0, 55.89473684210526, 57.78947368421052, 59.68421052631578, 61.57894736842104, 63.4736842105263, 65.36842105263156, 67.26315789473682, 69.15789473684211, 71.05263157894737, 72.94736842105263, 74.84210526315789, 76.73684210526315, 78.63157894736841, 80.52631578947367, 82.42105263157893, 84.31578947368419, 86.21052631578945, 88.10526315789474, 90.0], dtype='float64', name='lat')) - lonPandasIndex
PandasIndex(Index([ 0.0, 2.5, 5.0, 7.5, 10.0, 12.5, 15.0, 17.5, 20.0, 22.5, ... 335.0, 337.5, 340.0, 342.5, 345.0, 347.5, 350.0, 352.5, 355.0, 357.5], dtype='float64', name='lon', length=144)) - timePandasIndex
PandasIndex(CFTimeIndex([0001-01-17 00:00:00, 0001-02-14 00:00:00, 0001-03-17 00:00:00, 0001-04-16 00:00:00, 0001-05-17 00:00:00, 0001-06-16 00:00:00, 0001-07-17 00:00:00, 0001-08-17 00:00:00, 0001-09-16 00:00:00, 0001-10-17 00:00:00, 0001-11-16 00:00:00, 0001-12-17 00:00:00], dtype='object', length=12, calendar='noleap', freq=None))
- units :
- m/s
- long_name :
- Convective precipitation rate (liq + ice)
- cell_methods :
- time: mean
ds_PI.PRECL
<xarray.DataArray 'PRECL' (time: 12, lat: 96, lon: 144)> Size: 664kB
dask.array<concatenate, shape=(12, 96, 144), dtype=float32, chunksize=(1, 96, 144), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float64 768B -90.0 -88.11 -86.21 -84.32 ... 86.21 88.11 90.0
* lon (lon) float64 1kB 0.0 2.5 5.0 7.5 10.0 ... 350.0 352.5 355.0 357.5
* time (time) object 96B 0001-01-17 00:00:00 ... 0001-12-17 00:00:00
Attributes:
units: m/s
long_name: Large-scale (stable) precipitation rate (liq + ice)
cell_methods: time: mean- time: 12
- lat: 96
- lon: 144
- dask.array<chunksize=(1, 96, 144), meta=np.ndarray>
Array Chunk Bytes 648.00 kiB 54.00 kiB Shape (12, 96, 144) (1, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - lat(lat)float64-90.0 -88.11 -86.21 ... 88.11 90.0
- long_name :
- latitude
- units :
- degrees_north
array([-90. , -88.105263, -86.210526, -84.315789, -82.421053, -80.526316, -78.631579, -76.736842, -74.842105, -72.947368, -71.052632, -69.157895, -67.263158, -65.368421, -63.473684, -61.578947, -59.684211, -57.789474, -55.894737, -54. , -52.105263, -50.210526, -48.315789, -46.421053, -44.526316, -42.631579, -40.736842, -38.842105, -36.947368, -35.052632, -33.157895, -31.263158, -29.368421, -27.473684, -25.578947, -23.684211, -21.789474, -19.894737, -18. , -16.105263, -14.210526, -12.315789, -10.421053, -8.526316, -6.631579, -4.736842, -2.842105, -0.947368, 0.947368, 2.842105, 4.736842, 6.631579, 8.526316, 10.421053, 12.315789, 14.210526, 16.105263, 18. , 19.894737, 21.789474, 23.684211, 25.578947, 27.473684, 29.368421, 31.263158, 33.157895, 35.052632, 36.947368, 38.842105, 40.736842, 42.631579, 44.526316, 46.421053, 48.315789, 50.210526, 52.105263, 54. , 55.894737, 57.789474, 59.684211, 61.578947, 63.473684, 65.368421, 67.263158, 69.157895, 71.052632, 72.947368, 74.842105, 76.736842, 78.631579, 80.526316, 82.421053, 84.315789, 86.210526, 88.105263, 90. ]) - lon(lon)float640.0 2.5 5.0 ... 352.5 355.0 357.5
- long_name :
- longitude
- units :
- degrees_east
array([ 0. , 2.5, 5. , 7.5, 10. , 12.5, 15. , 17.5, 20. , 22.5, 25. , 27.5, 30. , 32.5, 35. , 37.5, 40. , 42.5, 45. , 47.5, 50. , 52.5, 55. , 57.5, 60. , 62.5, 65. , 67.5, 70. , 72.5, 75. , 77.5, 80. , 82.5, 85. , 87.5, 90. , 92.5, 95. , 97.5, 100. , 102.5, 105. , 107.5, 110. , 112.5, 115. , 117.5, 120. , 122.5, 125. , 127.5, 130. , 132.5, 135. , 137.5, 140. , 142.5, 145. , 147.5, 150. , 152.5, 155. , 157.5, 160. , 162.5, 165. , 167.5, 170. , 172.5, 175. , 177.5, 180. , 182.5, 185. , 187.5, 190. , 192.5, 195. , 197.5, 200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. , 332.5, 335. , 337.5, 340. , 342.5, 345. , 347.5, 350. , 352.5, 355. , 357.5]) - time(time)object0001-01-17 00:00:00 ... 0001-12-...
array([cftime.DatetimeNoLeap(1, 1, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 2, 14, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 3, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 4, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 5, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 6, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 7, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 8, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 9, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 10, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 12, 17, 0, 0, 0, 0, has_year_zero=True)], dtype=object)
- latPandasIndex
PandasIndex(Index([ -90.0, -88.10526315789474, -86.21052631578948, -84.3157894736842, -82.42105263157895, -80.52631578947368, -78.63157894736842, -76.73684210526316, -74.84210526315789, -72.94736842105263, -71.05263157894737, -69.15789473684211, -67.26315789473685, -65.36842105263158, -63.473684210526315, -61.578947368421055, -59.684210526315795, -57.78947368421053, -55.89473684210527, -54.0, -52.10526315789474, -50.21052631578947, -48.31578947368421, -46.42105263157895, -44.526315789473685, -42.631578947368425, -40.73684210526316, -38.8421052631579, -36.94736842105264, -35.05263157894737, -33.15789473684211, -31.263157894736842, -29.368421052631582, -27.473684210526322, -25.578947368421055, -23.684210526315795, -21.789473684210535, -19.89473684210526, -18.0, -16.10526315789474, -14.21052631578948, -12.31578947368422, -10.421052631578945, -8.526315789473685, -6.631578947368425, -4.736842105263165, -2.8421052631579045, -0.9473684210526301, 0.9473684210526301, 2.8421052631578902, 4.73684210526315, 6.631578947368411, 8.526315789473685, 10.421052631578945, 12.315789473684205, 14.210526315789465, 16.105263157894726, 18.0, 19.89473684210526, 21.78947368421052, 23.68421052631578, 25.57894736842104, 27.473684210526315, 29.368421052631575, 31.263157894736835, 33.157894736842096, 35.052631578947356, 36.94736842105263, 38.84210526315789, 40.73684210526315, 42.63157894736841, 44.52631578947367, 46.42105263157893, 48.31578947368419, 50.21052631578948, 52.10526315789474, 54.0, 55.89473684210526, 57.78947368421052, 59.68421052631578, 61.57894736842104, 63.4736842105263, 65.36842105263156, 67.26315789473682, 69.15789473684211, 71.05263157894737, 72.94736842105263, 74.84210526315789, 76.73684210526315, 78.63157894736841, 80.52631578947367, 82.42105263157893, 84.31578947368419, 86.21052631578945, 88.10526315789474, 90.0], dtype='float64', name='lat')) - lonPandasIndex
PandasIndex(Index([ 0.0, 2.5, 5.0, 7.5, 10.0, 12.5, 15.0, 17.5, 20.0, 22.5, ... 335.0, 337.5, 340.0, 342.5, 345.0, 347.5, 350.0, 352.5, 355.0, 357.5], dtype='float64', name='lon', length=144)) - timePandasIndex
PandasIndex(CFTimeIndex([0001-01-17 00:00:00, 0001-02-14 00:00:00, 0001-03-17 00:00:00, 0001-04-16 00:00:00, 0001-05-17 00:00:00, 0001-06-16 00:00:00, 0001-07-17 00:00:00, 0001-08-17 00:00:00, 0001-09-16 00:00:00, 0001-10-17 00:00:00, 0001-11-16 00:00:00, 0001-12-17 00:00:00], dtype='object', length=12, calendar='noleap', freq=None))
- units :
- m/s
- long_name :
- Large-scale (stable) precipitation rate (liq + ice)
- cell_methods :
- time: mean
m_p_s_to_mm_p_day = 86400000
prec_PI = (ds_PI.PRECC + ds_PI.PRECL).isel(time=slice(5, 8)).mean('time') * m_p_s_to_mm_p_day
prec_MH = (ds_MH.PRECC + ds_MH.PRECL).isel(time=slice(5, 8)).mean('time') * m_p_s_to_mm_p_day
prec_PI
<xarray.DataArray (lat: 96, lon: 144)> Size: 111kB dask.array<mul, shape=(96, 144), dtype=float64, chunksize=(96, 144), chunktype=numpy.ndarray> Coordinates: * lat (lat) float64 768B -90.0 -88.11 -86.21 -84.32 ... 86.21 88.11 90.0 * lon (lon) float64 1kB 0.0 2.5 5.0 7.5 10.0 ... 350.0 352.5 355.0 357.5
- lat: 96
- lon: 144
- dask.array<chunksize=(96, 144), meta=np.ndarray>
Array Chunk Bytes 108.00 kiB 108.00 kiB Shape (96, 144) (96, 144) Dask graph 1 chunks in 55 graph layers Data type float64 numpy.ndarray - lat(lat)float64-90.0 -88.11 -86.21 ... 88.11 90.0
- long_name :
- latitude
- units :
- degrees_north
array([-90. , -88.105263, -86.210526, -84.315789, -82.421053, -80.526316, -78.631579, -76.736842, -74.842105, -72.947368, -71.052632, -69.157895, -67.263158, -65.368421, -63.473684, -61.578947, -59.684211, -57.789474, -55.894737, -54. , -52.105263, -50.210526, -48.315789, -46.421053, -44.526316, -42.631579, -40.736842, -38.842105, -36.947368, -35.052632, -33.157895, -31.263158, -29.368421, -27.473684, -25.578947, -23.684211, -21.789474, -19.894737, -18. , -16.105263, -14.210526, -12.315789, -10.421053, -8.526316, -6.631579, -4.736842, -2.842105, -0.947368, 0.947368, 2.842105, 4.736842, 6.631579, 8.526316, 10.421053, 12.315789, 14.210526, 16.105263, 18. , 19.894737, 21.789474, 23.684211, 25.578947, 27.473684, 29.368421, 31.263158, 33.157895, 35.052632, 36.947368, 38.842105, 40.736842, 42.631579, 44.526316, 46.421053, 48.315789, 50.210526, 52.105263, 54. , 55.894737, 57.789474, 59.684211, 61.578947, 63.473684, 65.368421, 67.263158, 69.157895, 71.052632, 72.947368, 74.842105, 76.736842, 78.631579, 80.526316, 82.421053, 84.315789, 86.210526, 88.105263, 90. ]) - lon(lon)float640.0 2.5 5.0 ... 352.5 355.0 357.5
- long_name :
- longitude
- units :
- degrees_east
array([ 0. , 2.5, 5. , 7.5, 10. , 12.5, 15. , 17.5, 20. , 22.5, 25. , 27.5, 30. , 32.5, 35. , 37.5, 40. , 42.5, 45. , 47.5, 50. , 52.5, 55. , 57.5, 60. , 62.5, 65. , 67.5, 70. , 72.5, 75. , 77.5, 80. , 82.5, 85. , 87.5, 90. , 92.5, 95. , 97.5, 100. , 102.5, 105. , 107.5, 110. , 112.5, 115. , 117.5, 120. , 122.5, 125. , 127.5, 130. , 132.5, 135. , 137.5, 140. , 142.5, 145. , 147.5, 150. , 152.5, 155. , 157.5, 160. , 162.5, 165. , 167.5, 170. , 172.5, 175. , 177.5, 180. , 182.5, 185. , 187.5, 190. , 192.5, 195. , 197.5, 200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. , 332.5, 335. , 337.5, 340. , 342.5, 345. , 347.5, 350. , 352.5, 355. , 357.5])
- latPandasIndex
PandasIndex(Index([ -90.0, -88.10526315789474, -86.21052631578948, -84.3157894736842, -82.42105263157895, -80.52631578947368, -78.63157894736842, -76.73684210526316, -74.84210526315789, -72.94736842105263, -71.05263157894737, -69.15789473684211, -67.26315789473685, -65.36842105263158, -63.473684210526315, -61.578947368421055, -59.684210526315795, -57.78947368421053, -55.89473684210527, -54.0, -52.10526315789474, -50.21052631578947, -48.31578947368421, -46.42105263157895, -44.526315789473685, -42.631578947368425, -40.73684210526316, -38.8421052631579, -36.94736842105264, -35.05263157894737, -33.15789473684211, -31.263157894736842, -29.368421052631582, -27.473684210526322, -25.578947368421055, -23.684210526315795, -21.789473684210535, -19.89473684210526, -18.0, -16.10526315789474, -14.21052631578948, -12.31578947368422, -10.421052631578945, -8.526315789473685, -6.631578947368425, -4.736842105263165, -2.8421052631579045, -0.9473684210526301, 0.9473684210526301, 2.8421052631578902, 4.73684210526315, 6.631578947368411, 8.526315789473685, 10.421052631578945, 12.315789473684205, 14.210526315789465, 16.105263157894726, 18.0, 19.89473684210526, 21.78947368421052, 23.68421052631578, 25.57894736842104, 27.473684210526315, 29.368421052631575, 31.263157894736835, 33.157894736842096, 35.052631578947356, 36.94736842105263, 38.84210526315789, 40.73684210526315, 42.63157894736841, 44.52631578947367, 46.42105263157893, 48.31578947368419, 50.21052631578948, 52.10526315789474, 54.0, 55.89473684210526, 57.78947368421052, 59.68421052631578, 61.57894736842104, 63.4736842105263, 65.36842105263156, 67.26315789473682, 69.15789473684211, 71.05263157894737, 72.94736842105263, 74.84210526315789, 76.73684210526315, 78.63157894736841, 80.52631578947367, 82.42105263157893, 84.31578947368419, 86.21052631578945, 88.10526315789474, 90.0], dtype='float64', name='lat')) - lonPandasIndex
PandasIndex(Index([ 0.0, 2.5, 5.0, 7.5, 10.0, 12.5, 15.0, 17.5, 20.0, 22.5, ... 335.0, 337.5, 340.0, 342.5, 345.0, 347.5, 350.0, 352.5, 355.0, 357.5], dtype='float64', name='lon', length=144))
Plot total precipitation of piControl#
fig, ax = plt.subplots(figsize=(3, 1.5), subplot_kw={
'projection': ccrs.Robinson(central_longitude=210)})
p1 = ax.contourf(prec_PI.lon, prec_PI.lat, prec_PI,
cmap='YlGnBu',
levels=np.linspace(0, 17, 18),
extend='both',
transform=ccrs.PlateCarree())
ax.coastlines(linewidth=0.5)
plt.colorbar(p1)
ax.set_title("PI precipitation")
Text(0.5, 1.0, 'PI precipitation')
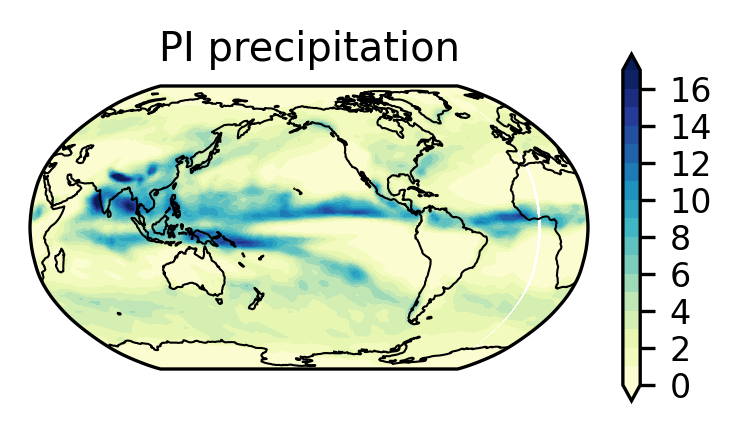
Let’s use add_cyclic_point to get rid of the “white strip” in the plot#
fig, ax = plt.subplots(figsize=(3, 1.5), subplot_kw={
'projection': ccrs.Robinson(central_longitude=210)})
# Note the differences in the next two lines
prec_PI_new, lon_new = add_cyclic_point(prec_PI, prec_PI.lon)
p1 = ax.contourf(lon_new, prec_PI.lat, prec_PI_new,
cmap='YlGnBu',
levels=np.linspace(0, 17, 18),
extend='both',
transform=ccrs.PlateCarree())
ax.coastlines(linewidth=0.5)
plt.colorbar(p1)
ax.set_title("PI precipitation")
Text(0.5, 1.0, 'PI precipitation')
Add your own plot of the midHolocene precipitatin#
fig, ax = plt.subplots(figsize=(3, 1.5), subplot_kw={
'projection': ccrs.Robinson(central_longitude=210)})

Add your own plot of the midHolocene - piControl#
fig, ax = plt.subplots(figsize=(3, 1.5), subplot_kw={
'projection': ccrs.Robinson(central_longitude=210)})

Click here for the solution
Copy and paste the code into the above cell
prec_MH_new, lon_new = add_cyclic_point(prec_MH, prec_MH.lon)
p1 = ax.contourf(lon_new, prec_MH.lat, prec_MH_new,
cmap='YlGnBu',
levels=np.linspace(0, 17, 18),
extend='both',
transform=ccrs.PlateCarree())
ax.coastlines(linewidth=0.5)
plt.colorbar(p1)
ax.set_title("MH precipitation")
p1 = ax.contourf(lon_new, prec_MH.lat, prec_MH_new - prec_PI_new,
cmap='BrBG',
levels=np.linspace(-5, 5, 21),
extend='both',
transform=ccrs.PlateCarree())
ax.coastlines(linewidth=0.5)
plt.colorbar(p1)
ax.set_title("MH precipitation anomalies")
Discussion#
Do you see the ITCZ in the piControl?
How does the ITCZ change in the mid-Holocene?
Do you see changes of the monsoon precipitation? Is it consistent with findings from Kutzbach and Otto-Bliesner (1981, 1982)?
Ask Kathleen, Jane, Daniel, and Kevin about ITCZ and monsoon!
Analysis 4: how does the MH orbital forcing impact the atmospheric circulation?#
We plot the zonal mean zonal wind of PI in the Northern Hemisphere summer (JJA)
We use
plt.gca().invert_yaxis()to invert the y-axis such that the high pressure is at the bottom
ds_PI.U
<xarray.DataArray 'U' (time: 12, lev: 26, lat: 96, lon: 144)> Size: 17MB
dask.array<concatenate, shape=(12, 26, 96, 144), dtype=float32, chunksize=(1, 26, 96, 144), chunktype=numpy.ndarray>
Coordinates:
* lat (lat) float64 768B -90.0 -88.11 -86.21 -84.32 ... 86.21 88.11 90.0
* lon (lon) float64 1kB 0.0 2.5 5.0 7.5 10.0 ... 350.0 352.5 355.0 357.5
* lev (lev) float64 208B 3.545 7.389 13.97 23.94 ... 929.6 970.6 992.6
* time (time) object 96B 0001-01-17 00:00:00 ... 0001-12-17 00:00:00
Attributes:
mdims: 1
units: m/s
long_name: Zonal wind
cell_methods: time: mean- time: 12
- lev: 26
- lat: 96
- lon: 144
- dask.array<chunksize=(1, 26, 96, 144), meta=np.ndarray>
Array Chunk Bytes 16.45 MiB 1.37 MiB Shape (12, 26, 96, 144) (1, 26, 96, 144) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - lat(lat)float64-90.0 -88.11 -86.21 ... 88.11 90.0
- long_name :
- latitude
- units :
- degrees_north
array([-90. , -88.105263, -86.210526, -84.315789, -82.421053, -80.526316, -78.631579, -76.736842, -74.842105, -72.947368, -71.052632, -69.157895, -67.263158, -65.368421, -63.473684, -61.578947, -59.684211, -57.789474, -55.894737, -54. , -52.105263, -50.210526, -48.315789, -46.421053, -44.526316, -42.631579, -40.736842, -38.842105, -36.947368, -35.052632, -33.157895, -31.263158, -29.368421, -27.473684, -25.578947, -23.684211, -21.789474, -19.894737, -18. , -16.105263, -14.210526, -12.315789, -10.421053, -8.526316, -6.631579, -4.736842, -2.842105, -0.947368, 0.947368, 2.842105, 4.736842, 6.631579, 8.526316, 10.421053, 12.315789, 14.210526, 16.105263, 18. , 19.894737, 21.789474, 23.684211, 25.578947, 27.473684, 29.368421, 31.263158, 33.157895, 35.052632, 36.947368, 38.842105, 40.736842, 42.631579, 44.526316, 46.421053, 48.315789, 50.210526, 52.105263, 54. , 55.894737, 57.789474, 59.684211, 61.578947, 63.473684, 65.368421, 67.263158, 69.157895, 71.052632, 72.947368, 74.842105, 76.736842, 78.631579, 80.526316, 82.421053, 84.315789, 86.210526, 88.105263, 90. ]) - lon(lon)float640.0 2.5 5.0 ... 352.5 355.0 357.5
- long_name :
- longitude
- units :
- degrees_east
array([ 0. , 2.5, 5. , 7.5, 10. , 12.5, 15. , 17.5, 20. , 22.5, 25. , 27.5, 30. , 32.5, 35. , 37.5, 40. , 42.5, 45. , 47.5, 50. , 52.5, 55. , 57.5, 60. , 62.5, 65. , 67.5, 70. , 72.5, 75. , 77.5, 80. , 82.5, 85. , 87.5, 90. , 92.5, 95. , 97.5, 100. , 102.5, 105. , 107.5, 110. , 112.5, 115. , 117.5, 120. , 122.5, 125. , 127.5, 130. , 132.5, 135. , 137.5, 140. , 142.5, 145. , 147.5, 150. , 152.5, 155. , 157.5, 160. , 162.5, 165. , 167.5, 170. , 172.5, 175. , 177.5, 180. , 182.5, 185. , 187.5, 190. , 192.5, 195. , 197.5, 200. , 202.5, 205. , 207.5, 210. , 212.5, 215. , 217.5, 220. , 222.5, 225. , 227.5, 230. , 232.5, 235. , 237.5, 240. , 242.5, 245. , 247.5, 250. , 252.5, 255. , 257.5, 260. , 262.5, 265. , 267.5, 270. , 272.5, 275. , 277.5, 280. , 282.5, 285. , 287.5, 290. , 292.5, 295. , 297.5, 300. , 302.5, 305. , 307.5, 310. , 312.5, 315. , 317.5, 320. , 322.5, 325. , 327.5, 330. , 332.5, 335. , 337.5, 340. , 342.5, 345. , 347.5, 350. , 352.5, 355. , 357.5]) - lev(lev)float643.545 7.389 13.97 ... 970.6 992.6
- long_name :
- hybrid level at midpoints (1000*(A+B))
- units :
- hPa
- positive :
- down
- standard_name :
- atmosphere_hybrid_sigma_pressure_coordinate
- formula_terms :
- a: hyam b: hybm p0: P0 ps: PS
array([ 3.544638, 7.388814, 13.967214, 23.944625, 37.23029 , 53.114605, 70.05915 , 85.439115, 100.514695, 118.250335, 139.115395, 163.66207 , 192.539935, 226.513265, 266.481155, 313.501265, 368.81798 , 433.895225, 510.455255, 600.5242 , 696.79629 , 787.70206 , 867.16076 , 929.648875, 970.55483 , 992.5561 ]) - time(time)object0001-01-17 00:00:00 ... 0001-12-...
array([cftime.DatetimeNoLeap(1, 1, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 2, 14, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 3, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 4, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 5, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 6, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 7, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 8, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 9, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 10, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 12, 17, 0, 0, 0, 0, has_year_zero=True)], dtype=object)
- latPandasIndex
PandasIndex(Index([ -90.0, -88.10526315789474, -86.21052631578948, -84.3157894736842, -82.42105263157895, -80.52631578947368, -78.63157894736842, -76.73684210526316, -74.84210526315789, -72.94736842105263, -71.05263157894737, -69.15789473684211, -67.26315789473685, -65.36842105263158, -63.473684210526315, -61.578947368421055, -59.684210526315795, -57.78947368421053, -55.89473684210527, -54.0, -52.10526315789474, -50.21052631578947, -48.31578947368421, -46.42105263157895, -44.526315789473685, -42.631578947368425, -40.73684210526316, -38.8421052631579, -36.94736842105264, -35.05263157894737, -33.15789473684211, -31.263157894736842, -29.368421052631582, -27.473684210526322, -25.578947368421055, -23.684210526315795, -21.789473684210535, -19.89473684210526, -18.0, -16.10526315789474, -14.21052631578948, -12.31578947368422, -10.421052631578945, -8.526315789473685, -6.631578947368425, -4.736842105263165, -2.8421052631579045, -0.9473684210526301, 0.9473684210526301, 2.8421052631578902, 4.73684210526315, 6.631578947368411, 8.526315789473685, 10.421052631578945, 12.315789473684205, 14.210526315789465, 16.105263157894726, 18.0, 19.89473684210526, 21.78947368421052, 23.68421052631578, 25.57894736842104, 27.473684210526315, 29.368421052631575, 31.263157894736835, 33.157894736842096, 35.052631578947356, 36.94736842105263, 38.84210526315789, 40.73684210526315, 42.63157894736841, 44.52631578947367, 46.42105263157893, 48.31578947368419, 50.21052631578948, 52.10526315789474, 54.0, 55.89473684210526, 57.78947368421052, 59.68421052631578, 61.57894736842104, 63.4736842105263, 65.36842105263156, 67.26315789473682, 69.15789473684211, 71.05263157894737, 72.94736842105263, 74.84210526315789, 76.73684210526315, 78.63157894736841, 80.52631578947367, 82.42105263157893, 84.31578947368419, 86.21052631578945, 88.10526315789474, 90.0], dtype='float64', name='lat')) - lonPandasIndex
PandasIndex(Index([ 0.0, 2.5, 5.0, 7.5, 10.0, 12.5, 15.0, 17.5, 20.0, 22.5, ... 335.0, 337.5, 340.0, 342.5, 345.0, 347.5, 350.0, 352.5, 355.0, 357.5], dtype='float64', name='lon', length=144)) - levPandasIndex
PandasIndex(Index([3.5446380000000097, 7.3888135000000075, 13.967214000000006, 23.944625, 37.23029000000011, 53.1146050000002, 70.05915000000029, 85.43911500000031, 100.51469500000029, 118.25033500000026, 139.11539500000046, 163.66207000000043, 192.53993500000033, 226.51326500000036, 266.4811550000001, 313.5012650000006, 368.81798000000157, 433.8952250000011, 510.45525500000167, 600.5242000000027, 696.7962900000033, 787.7020600000026, 867.1607600000013, 929.6488750000024, 970.5548300000014, 992.5560999999998], dtype='float64', name='lev')) - timePandasIndex
PandasIndex(CFTimeIndex([0001-01-17 00:00:00, 0001-02-14 00:00:00, 0001-03-17 00:00:00, 0001-04-16 00:00:00, 0001-05-17 00:00:00, 0001-06-16 00:00:00, 0001-07-17 00:00:00, 0001-08-17 00:00:00, 0001-09-16 00:00:00, 0001-10-17 00:00:00, 0001-11-16 00:00:00, 0001-12-17 00:00:00], dtype='object', length=12, calendar='noleap', freq=None))
- mdims :
- 1
- units :
- m/s
- long_name :
- Zonal wind
- cell_methods :
- time: mean
Plot the zonal mean of u-winds of JJA#
U_PI_za_jja = ds_PI.U.isel(time=slice(5, 8)).mean(('lon', 'time'))
U_PI_za_jja.plot.contourf(figsize=(3, 1.5),
levels=np.linspace(-50, 50, 21), extend='both', )
plt.gca().invert_yaxis()
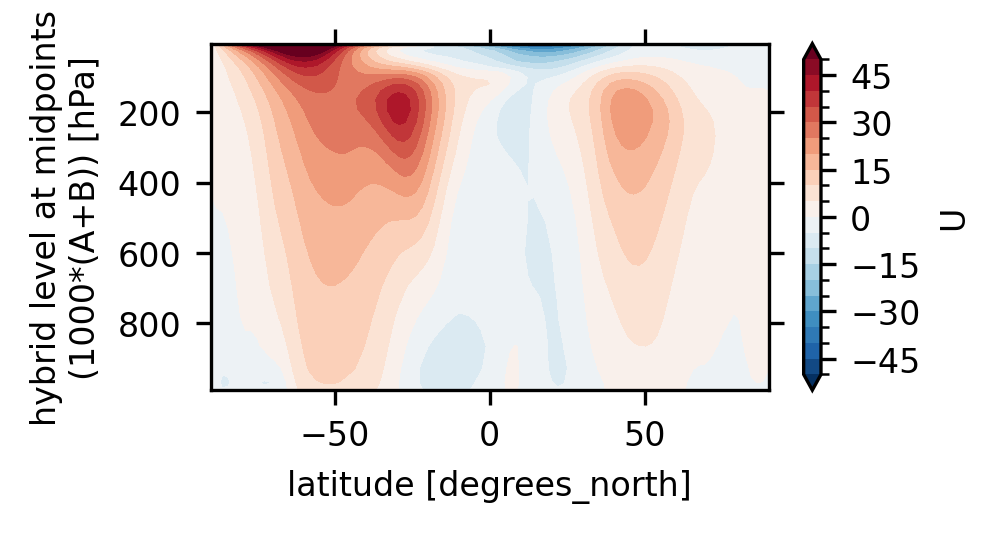
Add your own plot to show the changes during the midHolocene#
Click here for the solution
Copy and paste the code into the above cell
dU_MH_za_jja = (ds_MH.U - ds_PI.U).isel(time=slice(5, 8)).mean(('lon', 'time'))
dU_MH_za_jja.plot.contourf(figsize=(3, 1.5),
levels=np.linspace(-10, 10, 21), extend='both', )
plt.gca().invert_yaxis()
Did you notice any problem with the above plots?#
Check out the name of the vertical levels. it is called hybrid sigma-pressure coordinate, which is NOT the pressure coordinate.
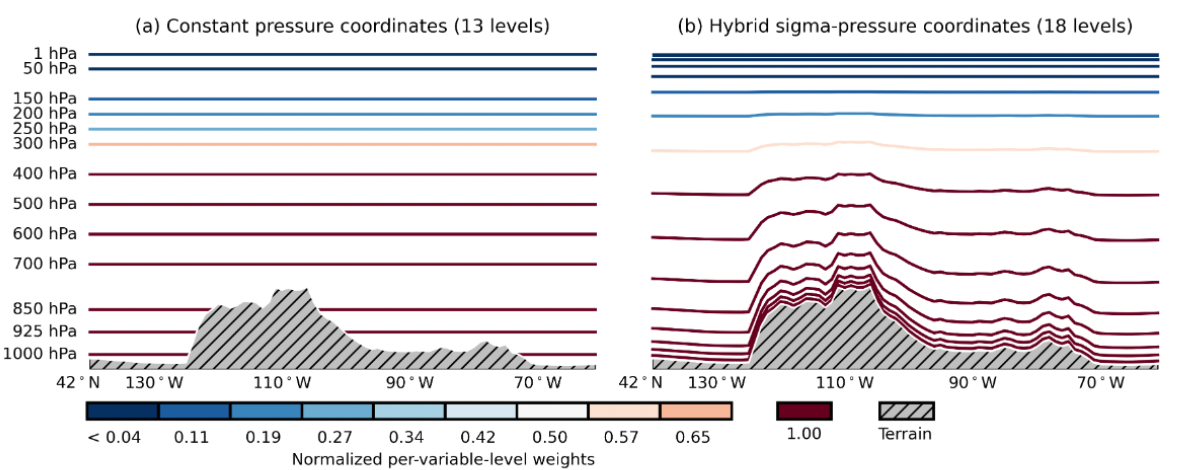
Figure: Atmopshere vertical coordinates
We need to interpolate from the hybrid sigma-pressure coordinate into the normal pressure coordinate
ds_PI.lev.standard_name
'atmosphere_hybrid_sigma_pressure_coordinate'
Use geocat interpolation to interpolate from hybrid sigma-pressure to presure coordinate#
P_new_mb = np.array([1, 10, 20, 50., 100., 200., 300.,
400., 500., 600., 700., 800., 900., 1000.])
p0_mb = 1000.
Up_PI = interpolation.interp_hybrid_to_pressure(
ds_PI.U,
ds_PI.PS,
ds_PI.hyam,
ds_PI.hybm,
p0=p0_mb*100.,
new_levels=P_new_mb*100.,
extrapolate=False)
Up_MH = interpolation.interp_hybrid_to_pressure(
ds_MH.U,
ds_MH.PS,
ds_MH.hyam,
ds_MH.hybm,
p0=p0_mb*100.,
new_levels=P_new_mb*100.,
extrapolate=False)
Plot the correct results!#
fig, axes = plt.subplots(nrows=1, ncols=2,
figsize=(8, 2),
constrained_layout=True)
Up_PI.isel(time=slice(5, 8)).mean(('lon', 'time')).plot.contourf(
ax=axes[0], levels=np.linspace(-50, 50, 21), extend='both')
(Up_MH - Up_PI).isel(time=slice(5, 8)).mean(('lon', 'time')).plot.contourf(
ax=axes[1], levels=np.linspace(-10, 10, 21), extend='both')
for ax in axes:
ax.invert_yaxis()
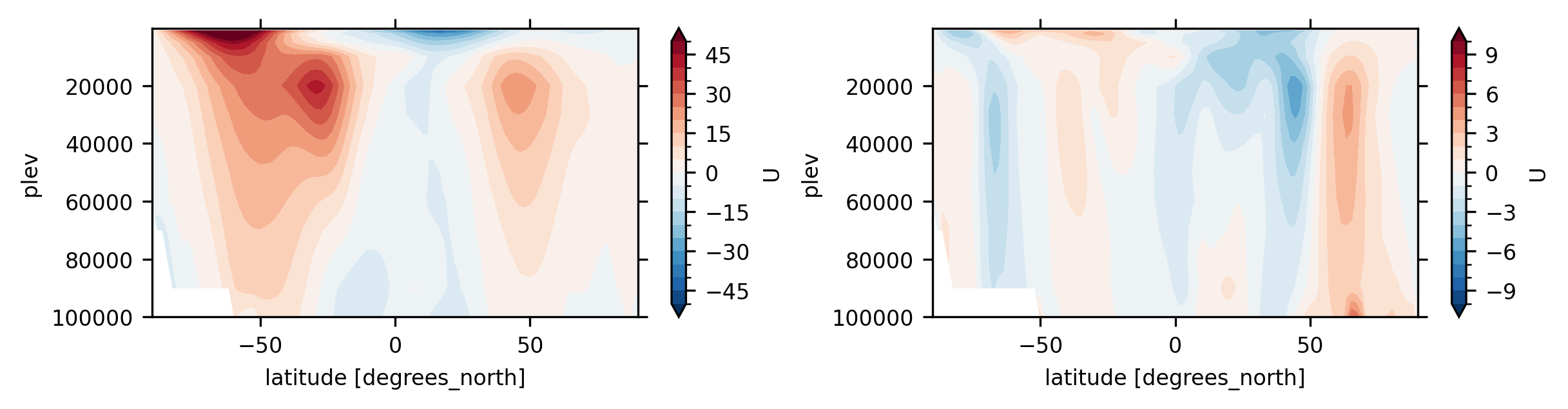
Small group discussion#
Which hemisphere has a stronger jet stream? Why? (remember that the plots are for JJA, the NH summer)
Does the mid-Holocene orbital forcing shift the jet stream?
Ask Sylvia, Jane and Kevin about the atmosphere circulation!
Analysis 5: how about sea-surface temperature?#
Ocean data is in
ocn/histSST is the top level of TEMP
hist_dir = '/ocn/hist/'
files_PI_ocn = glob.glob(storage_dir + case_PI + hist_dir + '*.pop.h.0001*')
files_MH_ocn = glob.glob(storage_dir + case_MH + hist_dir + '*.pop.h.0001*')
print(*files_PI_ocn, sep='\n')
print(*files_MH_ocn, sep='\n')
ds_PI_ocn = xr.open_mfdataset(files_PI_ocn)
ds_MH_ocn = xr.open_mfdataset(files_MH_ocn)
# Again, we need this fix to get the correct time, i.e., month 1 to month 12
ds_PI_ocn['time'] = ds_PI_ocn.time.get_index('time') - timedelta(days=15)
ds_MH_ocn['time'] = ds_MH_ocn.time.get_index('time') - timedelta(days=15)
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/ocn/hist/b.e21.B1850.f19_g17.piControl.001.pop.h.0001-03.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/ocn/hist/b.e21.B1850.f19_g17.piControl.001.pop.h.0001-01.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/ocn/hist/b.e21.B1850.f19_g17.piControl.001.pop.h.0001-02.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/ocn/hist/b.e21.B1850.f19_g17.piControl.001.pop.h.0001-09.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/ocn/hist/b.e21.B1850.f19_g17.piControl.001.pop.h.0001-10.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/ocn/hist/b.e21.B1850.f19_g17.piControl.001.pop.h.0001-08.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/ocn/hist/b.e21.B1850.f19_g17.piControl.001.pop.h.0001-11.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/ocn/hist/b.e21.B1850.f19_g17.piControl.001.pop.h.0001-07.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/ocn/hist/b.e21.B1850.f19_g17.piControl.001.pop.h.0001-05.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/ocn/hist/b.e21.B1850.f19_g17.piControl.001.pop.h.0001-06.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/ocn/hist/b.e21.B1850.f19_g17.piControl.001.pop.h.0001-12.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.piControl.001/ocn/hist/b.e21.B1850.f19_g17.piControl.001.pop.h.0001-04.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/ocn/hist/b.e21.B1850.f19_g17.midHolocene.001.pop.h.0001-05.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/ocn/hist/b.e21.B1850.f19_g17.midHolocene.001.pop.h.0001-11.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/ocn/hist/b.e21.B1850.f19_g17.midHolocene.001.pop.h.0001-01.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/ocn/hist/b.e21.B1850.f19_g17.midHolocene.001.pop.h.0001-02.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/ocn/hist/b.e21.B1850.f19_g17.midHolocene.001.pop.h.0001-08.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/ocn/hist/b.e21.B1850.f19_g17.midHolocene.001.pop.h.0001-10.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/ocn/hist/b.e21.B1850.f19_g17.midHolocene.001.pop.h.0001-04.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/ocn/hist/b.e21.B1850.f19_g17.midHolocene.001.pop.h.0001-06.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/ocn/hist/b.e21.B1850.f19_g17.midHolocene.001.pop.h.0001-07.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/ocn/hist/b.e21.B1850.f19_g17.midHolocene.001.pop.h.0001-09.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/ocn/hist/b.e21.B1850.f19_g17.midHolocene.001.pop.h.0001-03.nc
/glade/derecho/scratch/jiangzhu/archive/b.e21.B1850.f19_g17.midHolocene.001/ocn/hist/b.e21.B1850.f19_g17.midHolocene.001.pop.h.0001-12.nc
ds_PI_ocn
<xarray.Dataset> Size: 20GB
Dimensions: (time: 12, d2: 2, z_t: 60, z_w: 60, nlat: 384,
nlon: 320, z_w_bot: 60, z_w_top: 60,
transport_reg: 2, moc_comp: 3, moc_z: 61,
lat_aux_grid: 395, transport_comp: 5, z_t_150m: 15)
Coordinates: (12/15)
moc_components (moc_comp) |S384 1kB dask.array<chunksize=(3,), meta=np.ndarray>
transport_components (transport_comp) |S384 2kB dask.array<chunksize=(5,), meta=np.ndarray>
transport_regions (transport_reg) |S384 768B dask.array<chunksize=(2,), meta=np.ndarray>
* time (time) object 96B 0001-01-17 00:00:00 ... 0001-12...
* z_t (z_t) float32 240B 500.0 1.5e+03 ... 5.375e+05
* z_t_150m (z_t_150m) float32 60B 500.0 1.5e+03 ... 1.45e+04
... ...
* lat_aux_grid (lat_aux_grid) float32 2kB -79.49 -78.95 ... 90.0
* moc_z (moc_z) float32 244B 0.0 1e+03 ... 5.25e+05 5.5e+05
ULONG (nlat, nlon) float64 983kB dask.array<chunksize=(384, 320), meta=np.ndarray>
ULAT (nlat, nlon) float64 983kB dask.array<chunksize=(384, 320), meta=np.ndarray>
TLONG (nlat, nlon) float64 983kB dask.array<chunksize=(384, 320), meta=np.ndarray>
TLAT (nlat, nlon) float64 983kB dask.array<chunksize=(384, 320), meta=np.ndarray>
Dimensions without coordinates: d2, nlat, nlon, transport_reg, moc_comp,
transport_comp
Data variables: (12/171)
time_bound (time, d2) object 192B dask.array<chunksize=(1, 2), meta=np.ndarray>
dz (time, z_t) float32 3kB dask.array<chunksize=(1, 60), meta=np.ndarray>
dzw (time, z_w) float32 3kB dask.array<chunksize=(1, 60), meta=np.ndarray>
KMT (time, nlat, nlon) float64 12MB dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
KMU (time, nlat, nlon) float64 12MB dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
REGION_MASK (time, nlat, nlon) float64 12MB dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
... ...
XBLT (time, nlat, nlon) float32 6MB dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
TBLT (time, nlat, nlon) float32 6MB dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
BSF (time, nlat, nlon) float32 6MB dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
MOC (time, transport_reg, moc_comp, moc_z, lat_aux_grid) float32 7MB dask.array<chunksize=(1, 2, 3, 61, 395), meta=np.ndarray>
N_HEAT (time, transport_reg, transport_comp, lat_aux_grid) float32 190kB dask.array<chunksize=(1, 2, 5, 395), meta=np.ndarray>
N_SALT (time, transport_reg, transport_comp, lat_aux_grid) float32 190kB dask.array<chunksize=(1, 2, 5, 395), meta=np.ndarray>
Attributes:
title: b.e21.B1850.f19_g17.piControl.001
history: none
Conventions: CF-1.0; http://www.cgd.ucar.edu/cms/eaton/netcdf/CF-cu...
time_period_freq: month_1
model_doi_url: https://doi.org/10.5065/D67H1H0V
contents: Diagnostic and Prognostic Variables
source: CCSM POP2, the CCSM Ocean Component
revision: $Id$
calendar: All years have exactly 365 days.
start_time: This dataset was created on 2025-06-14 at 12:35:19.2
cell_methods: cell_methods = time: mean ==> the variable values are ...- time: 12
- d2: 2
- z_t: 60
- z_w: 60
- nlat: 384
- nlon: 320
- z_w_bot: 60
- z_w_top: 60
- transport_reg: 2
- moc_comp: 3
- moc_z: 61
- lat_aux_grid: 395
- transport_comp: 5
- z_t_150m: 15
- moc_components(moc_comp)|S384dask.array<chunksize=(3,), meta=np.ndarray>
- long_name :
- MOC component names
- units :
Array Chunk Bytes 1.12 kiB 1.12 kiB Shape (3,) (3,) Dask graph 1 chunks in 58 graph layers Data type |S384 numpy.ndarray - transport_components(transport_comp)|S384dask.array<chunksize=(5,), meta=np.ndarray>
- long_name :
- T,S transport components
- units :
Array Chunk Bytes 1.88 kiB 1.88 kiB Shape (5,) (5,) Dask graph 1 chunks in 58 graph layers Data type |S384 numpy.ndarray - transport_regions(transport_reg)|S384dask.array<chunksize=(2,), meta=np.ndarray>
- long_name :
- regions for all transport diagnostics
- units :
Array Chunk Bytes 768 B 768 B Shape (2,) (2,) Dask graph 1 chunks in 58 graph layers Data type |S384 numpy.ndarray - time(time)object0001-01-17 00:00:00 ... 0001-12-...
array([cftime.DatetimeNoLeap(1, 1, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 2, 14, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 3, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 4, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 5, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 6, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 7, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 8, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 9, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 10, 17, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 11, 16, 0, 0, 0, 0, has_year_zero=True), cftime.DatetimeNoLeap(1, 12, 17, 0, 0, 0, 0, has_year_zero=True)], dtype=object) - z_t(z_t)float32500.0 1.5e+03 ... 5.375e+05
- long_name :
- depth from surface to midpoint of layer
- units :
- centimeters
- positive :
- down
- valid_min :
- 500.0
- valid_max :
- 537500.0
array([5.000000e+02, 1.500000e+03, 2.500000e+03, 3.500000e+03, 4.500000e+03, 5.500000e+03, 6.500000e+03, 7.500000e+03, 8.500000e+03, 9.500000e+03, 1.050000e+04, 1.150000e+04, 1.250000e+04, 1.350000e+04, 1.450000e+04, 1.550000e+04, 1.650984e+04, 1.754790e+04, 1.862913e+04, 1.976603e+04, 2.097114e+04, 2.225783e+04, 2.364088e+04, 2.513702e+04, 2.676542e+04, 2.854837e+04, 3.051192e+04, 3.268680e+04, 3.510935e+04, 3.782276e+04, 4.087846e+04, 4.433777e+04, 4.827367e+04, 5.277280e+04, 5.793729e+04, 6.388626e+04, 7.075633e+04, 7.870025e+04, 8.788252e+04, 9.847059e+04, 1.106204e+05, 1.244567e+05, 1.400497e+05, 1.573946e+05, 1.764003e+05, 1.968944e+05, 2.186457e+05, 2.413972e+05, 2.649001e+05, 2.889385e+05, 3.133405e+05, 3.379793e+05, 3.627670e+05, 3.876452e+05, 4.125768e+05, 4.375392e+05, 4.625190e+05, 4.875083e+05, 5.125028e+05, 5.375000e+05], dtype=float32) - z_t_150m(z_t_150m)float32500.0 1.5e+03 ... 1.35e+04 1.45e+04
- long_name :
- depth from surface to midpoint of layer
- units :
- centimeters
- positive :
- down
- valid_min :
- 500.0
- valid_max :
- 14500.0
array([ 500., 1500., 2500., 3500., 4500., 5500., 6500., 7500., 8500., 9500., 10500., 11500., 12500., 13500., 14500.], dtype=float32) - z_w(z_w)float320.0 1e+03 2e+03 ... 5e+05 5.25e+05
- long_name :
- depth from surface to top of layer
- units :
- centimeters
- positive :
- down
- valid_min :
- 0.0
- valid_max :
- 525000.94
array([ 0. , 1000. , 2000. , 3000. , 4000. , 5000. , 6000. , 7000. , 8000. , 9000. , 10000. , 11000. , 12000. , 13000. , 14000. , 15000. , 16000. , 17019.682, 18076.129, 19182.125, 20349.932, 21592.344, 22923.312, 24358.453, 25915.58 , 27615.26 , 29481.47 , 31542.373, 33831.227, 36387.473, 39258.047, 42498.887, 46176.656, 50370.688, 55174.91 , 60699.668, 67072.86 , 74439.805, 82960.695, 92804.35 , 104136.82 , 117104.016, 131809.36 , 148290.08 , 166499.2 , 186301.44 , 207487.39 , 229803.9 , 252990.4 , 276809.84 , 301067.06 , 325613.84 , 350344.88 , 375189.2 , 400101.16 , 425052.47 , 450026.06 , 475012. , 500004.7 , 525000.94 ], dtype=float32) - z_w_top(z_w_top)float320.0 1e+03 2e+03 ... 5e+05 5.25e+05
- long_name :
- depth from surface to top of layer
- units :
- centimeters
- positive :
- down
- valid_min :
- 0.0
- valid_max :
- 525000.94
array([ 0. , 1000. , 2000. , 3000. , 4000. , 5000. , 6000. , 7000. , 8000. , 9000. , 10000. , 11000. , 12000. , 13000. , 14000. , 15000. , 16000. , 17019.682, 18076.129, 19182.125, 20349.932, 21592.344, 22923.312, 24358.453, 25915.58 , 27615.26 , 29481.47 , 31542.373, 33831.227, 36387.473, 39258.047, 42498.887, 46176.656, 50370.688, 55174.91 , 60699.668, 67072.86 , 74439.805, 82960.695, 92804.35 , 104136.82 , 117104.016, 131809.36 , 148290.08 , 166499.2 , 186301.44 , 207487.39 , 229803.9 , 252990.4 , 276809.84 , 301067.06 , 325613.84 , 350344.88 , 375189.2 , 400101.16 , 425052.47 , 450026.06 , 475012. , 500004.7 , 525000.94 ], dtype=float32) - z_w_bot(z_w_bot)float321e+03 2e+03 ... 5.25e+05 5.5e+05
- long_name :
- depth from surface to bottom of layer
- units :
- centimeters
- positive :
- down
- valid_min :
- 1000.0
- valid_max :
- 549999.06
array([ 1000. , 2000. , 3000. , 4000. , 5000. , 6000. , 7000. , 8000. , 9000. , 10000. , 11000. , 12000. , 13000. , 14000. , 15000. , 16000. , 17019.682, 18076.129, 19182.125, 20349.932, 21592.344, 22923.312, 24358.453, 25915.58 , 27615.26 , 29481.47 , 31542.373, 33831.227, 36387.473, 39258.047, 42498.887, 46176.656, 50370.688, 55174.91 , 60699.668, 67072.86 , 74439.805, 82960.695, 92804.35 , 104136.82 , 117104.016, 131809.36 , 148290.08 , 166499.2 , 186301.44 , 207487.39 , 229803.9 , 252990.4 , 276809.84 , 301067.06 , 325613.84 , 350344.88 , 375189.2 , 400101.16 , 425052.47 , 450026.06 , 475012. , 500004.7 , 525000.94 , 549999.06 ], dtype=float32) - lat_aux_grid(lat_aux_grid)float32-79.49 -78.95 -78.42 ... 89.47 90.0
- long_name :
- latitude grid for transport diagnostics
- units :
- degrees_north
- valid_min :
- -79.48815
- valid_max :
- 90.0
array([-79.48815 , -78.952896, -78.418655, ..., 88.948814, 89.47441 , 90. ], dtype=float32) - moc_z(moc_z)float320.0 1e+03 ... 5.25e+05 5.5e+05
- long_name :
- depth from surface to top of layer
- units :
- centimeters
- positive :
- down
- valid_min :
- 0.0
- valid_max :
- 549999.06
array([ 0. , 1000. , 2000. , 3000. , 4000. , 5000. , 6000. , 7000. , 8000. , 9000. , 10000. , 11000. , 12000. , 13000. , 14000. , 15000. , 16000. , 17019.682, 18076.129, 19182.125, 20349.932, 21592.344, 22923.312, 24358.453, 25915.58 , 27615.26 , 29481.47 , 31542.373, 33831.227, 36387.473, 39258.047, 42498.887, 46176.656, 50370.688, 55174.91 , 60699.668, 67072.86 , 74439.805, 82960.695, 92804.35 , 104136.82 , 117104.016, 131809.36 , 148290.08 , 166499.2 , 186301.44 , 207487.39 , 229803.9 , 252990.4 , 276809.84 , 301067.06 , 325613.84 , 350344.88 , 375189.2 , 400101.16 , 425052.47 , 450026.06 , 475012. , 500004.7 , 525000.94 , 549999.06 ], dtype=float32) - ULONG(nlat, nlon)float64dask.array<chunksize=(384, 320), meta=np.ndarray>
- long_name :
- array of u-grid longitudes
- units :
- degrees_east
Array Chunk Bytes 0.94 MiB 0.94 MiB Shape (384, 320) (384, 320) Dask graph 1 chunks in 55 graph layers Data type float64 numpy.ndarray - ULAT(nlat, nlon)float64dask.array<chunksize=(384, 320), meta=np.ndarray>
- long_name :
- array of u-grid latitudes
- units :
- degrees_north
Array Chunk Bytes 0.94 MiB 0.94 MiB Shape (384, 320) (384, 320) Dask graph 1 chunks in 55 graph layers Data type float64 numpy.ndarray - TLONG(nlat, nlon)float64dask.array<chunksize=(384, 320), meta=np.ndarray>
- long_name :
- array of t-grid longitudes
- units :
- degrees_east
Array Chunk Bytes 0.94 MiB 0.94 MiB Shape (384, 320) (384, 320) Dask graph 1 chunks in 55 graph layers Data type float64 numpy.ndarray - TLAT(nlat, nlon)float64dask.array<chunksize=(384, 320), meta=np.ndarray>
- long_name :
- array of t-grid latitudes
- units :
- degrees_north
Array Chunk Bytes 0.94 MiB 0.94 MiB Shape (384, 320) (384, 320) Dask graph 1 chunks in 55 graph layers Data type float64 numpy.ndarray
- time_bound(time, d2)objectdask.array<chunksize=(1, 2), meta=np.ndarray>
- long_name :
- boundaries for time-averaging interval
Array Chunk Bytes 192 B 16 B Shape (12, 2) (1, 2) Dask graph 12 chunks in 25 graph layers Data type object numpy.ndarray - dz(time, z_t)float32dask.array<chunksize=(1, 60), meta=np.ndarray>
- long_name :
- thickness of layer k
- units :
- centimeters
Array Chunk Bytes 2.81 kiB 240 B Shape (12, 60) (1, 60) Dask graph 12 chunks in 37 graph layers Data type float32 numpy.ndarray - dzw(time, z_w)float32dask.array<chunksize=(1, 60), meta=np.ndarray>
- long_name :
- midpoint of k to midpoint of k+1
- units :
- centimeters
Array Chunk Bytes 2.81 kiB 240 B Shape (12, 60) (1, 60) Dask graph 12 chunks in 37 graph layers Data type float32 numpy.ndarray - KMT(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- k Index of Deepest Grid Cell on T Grid
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - KMU(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- k Index of Deepest Grid Cell on U Grid
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - REGION_MASK(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- basin index number (signed integers)
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - UAREA(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- area of U cells
- units :
- centimeter^2
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - TAREA(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- area of T cells
- units :
- centimeter^2
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - HU(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- ocean depth at U points
- units :
- centimeter
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - HT(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- ocean depth at T points
- units :
- centimeter
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - DXU(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- x-spacing centered at U points
- units :
- centimeters
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - DYU(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- y-spacing centered at U points
- units :
- centimeters
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - DXT(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- x-spacing centered at T points
- units :
- centimeters
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - DYT(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- y-spacing centered at T points
- units :
- centimeters
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - HTN(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- cell widths on North sides of T cell
- units :
- centimeters
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - HTE(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- cell widths on East sides of T cell
- units :
- centimeters
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - HUS(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- cell widths on South sides of U cell
- units :
- centimeters
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - HUW(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- cell widths on West sides of U cell
- units :
- centimeters
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - ANGLE(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- angle grid makes with latitude line
- units :
- radians
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - ANGLET(time, nlat, nlon)float64dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- angle grid makes with latitude line on T grid
- units :
- radians
Array Chunk Bytes 11.25 MiB 0.94 MiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 37 graph layers Data type float64 numpy.ndarray - days_in_norm_year(time)timedelta64[ns]365 days 365 days ... 365 days
- long_name :
- Calendar Length
array([31536000000000000, 31536000000000000, 31536000000000000, 31536000000000000, 31536000000000000, 31536000000000000, 31536000000000000, 31536000000000000, 31536000000000000, 31536000000000000, 31536000000000000, 31536000000000000], dtype='timedelta64[ns]') - grav(time)float64980.6 980.6 980.6 ... 980.6 980.6
- long_name :
- Acceleration Due to Gravity
- units :
- centimeter/s^2
array([980.616, 980.616, 980.616, 980.616, 980.616, 980.616, 980.616, 980.616, 980.616, 980.616, 980.616, 980.616]) - omega(time)float647.292e-05 7.292e-05 ... 7.292e-05
- long_name :
- Earths Angular Velocity
- units :
- 1/second
array([7.29212352e-05, 7.29212352e-05, 7.29212352e-05, 7.29212352e-05, 7.29212352e-05, 7.29212352e-05, 7.29212352e-05, 7.29212352e-05, 7.29212352e-05, 7.29212352e-05, 7.29212352e-05, 7.29212352e-05]) - radius(time)float646.371e+08 6.371e+08 ... 6.371e+08
- long_name :
- Earths Radius
- units :
- centimeters
array([6.37122e+08, 6.37122e+08, 6.37122e+08, 6.37122e+08, 6.37122e+08, 6.37122e+08, 6.37122e+08, 6.37122e+08, 6.37122e+08, 6.37122e+08, 6.37122e+08, 6.37122e+08]) - cp_sw(time)float643.996e+07 3.996e+07 ... 3.996e+07
- long_name :
- Specific Heat of Sea Water
- units :
- erg/g/K
array([39960000., 39960000., 39960000., 39960000., 39960000., 39960000., 39960000., 39960000., 39960000., 39960000., 39960000., 39960000.]) - sound(time)float641.5e+05 1.5e+05 ... 1.5e+05 1.5e+05
- long_name :
- Speed of Sound
- units :
- centimeter/s
array([150000., 150000., 150000., 150000., 150000., 150000., 150000., 150000., 150000., 150000., 150000., 150000.]) - vonkar(time)float640.4 0.4 0.4 0.4 ... 0.4 0.4 0.4 0.4
- long_name :
- von Karman Constant
array([0.4, 0.4, 0.4, 0.4, 0.4, 0.4, 0.4, 0.4, 0.4, 0.4, 0.4, 0.4])
- cp_air(time)float641.005e+03 1.005e+03 ... 1.005e+03
- long_name :
- Heat Capacity of Air
- units :
- joule/kg/degK
array([1004.64, 1004.64, 1004.64, 1004.64, 1004.64, 1004.64, 1004.64, 1004.64, 1004.64, 1004.64, 1004.64, 1004.64]) - rho_air(time)float641.292 1.292 1.292 ... 1.292 1.292
- long_name :
- Ambient Air Density
- units :
- kg/m^3
array([1.29231828, 1.29231828, 1.29231828, 1.29231828, 1.29231828, 1.29231828, 1.29231828, 1.29231828, 1.29231828, 1.29231828, 1.29231828, 1.29231828]) - rho_sw(time)float641.026 1.026 1.026 ... 1.026 1.026
- long_name :
- Density of Sea Water
- units :
- gram/centimeter^3
array([1.026, 1.026, 1.026, 1.026, 1.026, 1.026, 1.026, 1.026, 1.026, 1.026, 1.026, 1.026]) - rho_fw(time)float641.0 1.0 1.0 1.0 ... 1.0 1.0 1.0 1.0
- long_name :
- Density of Fresh Water
- units :
- gram/centimeter^3
array([1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.])
- stefan_boltzmann(time)float645.67e-08 5.67e-08 ... 5.67e-08
- long_name :
- Stefan-Boltzmann Constant
- units :
- watt/m^2/degK^4
array([5.67e-08, 5.67e-08, 5.67e-08, 5.67e-08, 5.67e-08, 5.67e-08, 5.67e-08, 5.67e-08, 5.67e-08, 5.67e-08, 5.67e-08, 5.67e-08]) - latent_heat_vapor(time)float642.501e+06 2.501e+06 ... 2.501e+06
- long_name :
- Latent Heat of Vaporization
- units :
- J/kg
array([2501000., 2501000., 2501000., 2501000., 2501000., 2501000., 2501000., 2501000., 2501000., 2501000., 2501000., 2501000.]) - latent_heat_fusion(time)float643.337e+09 3.337e+09 ... 3.337e+09
- long_name :
- Latent Heat of Fusion
- units :
- erg/g
array([3.337e+09, 3.337e+09, 3.337e+09, 3.337e+09, 3.337e+09, 3.337e+09, 3.337e+09, 3.337e+09, 3.337e+09, 3.337e+09, 3.337e+09, 3.337e+09]) - latent_heat_fusion_mks(time)float643.337e+05 3.337e+05 ... 3.337e+05
- long_name :
- Latent Heat of Fusion
- units :
- J/kg
array([333700., 333700., 333700., 333700., 333700., 333700., 333700., 333700., 333700., 333700., 333700., 333700.]) - ocn_ref_salinity(time)float6434.7 34.7 34.7 ... 34.7 34.7 34.7
- long_name :
- Ocean Reference Salinity
- units :
- g/kg
array([34.7, 34.7, 34.7, 34.7, 34.7, 34.7, 34.7, 34.7, 34.7, 34.7, 34.7, 34.7]) - sea_ice_salinity(time)float644.0 4.0 4.0 4.0 ... 4.0 4.0 4.0 4.0
- long_name :
- Salinity of Sea Ice
- units :
- g/kg
array([4., 4., 4., 4., 4., 4., 4., 4., 4., 4., 4., 4.])
- T0_Kelvin(time)float64273.1 273.1 273.1 ... 273.1 273.1
- long_name :
- Zero Point for Celsius
- units :
- degK
array([273.15, 273.15, 273.15, 273.15, 273.15, 273.15, 273.15, 273.15, 273.15, 273.15, 273.15, 273.15]) - salt_to_ppt(time)float641e+03 1e+03 1e+03 ... 1e+03 1e+03
- long_name :
- Convert Salt in gram/gram to g/kg
array([1000., 1000., 1000., 1000., 1000., 1000., 1000., 1000., 1000., 1000., 1000., 1000.]) - ppt_to_salt(time)float640.001 0.001 0.001 ... 0.001 0.001
- long_name :
- Convert Salt in g/kg to gram/gram
array([0.001, 0.001, 0.001, 0.001, 0.001, 0.001, 0.001, 0.001, 0.001, 0.001, 0.001, 0.001]) - mass_to_Sv(time)float641e-12 1e-12 1e-12 ... 1e-12 1e-12
- long_name :
- Convert Mass Flux to Sverdrups
array([1.e-12, 1.e-12, 1.e-12, 1.e-12, 1.e-12, 1.e-12, 1.e-12, 1.e-12, 1.e-12, 1.e-12, 1.e-12, 1.e-12]) - heat_to_PW(time)float644.186e-15 4.186e-15 ... 4.186e-15
- long_name :
- Convert Heat Flux to Petawatts
array([4.186e-15, 4.186e-15, 4.186e-15, 4.186e-15, 4.186e-15, 4.186e-15, 4.186e-15, 4.186e-15, 4.186e-15, 4.186e-15, 4.186e-15, 4.186e-15]) - salt_to_Svppt(time)float641e-09 1e-09 1e-09 ... 1e-09 1e-09
- long_name :
- Convert Salt Flux to Sverdrups*g/kg
array([1.e-09, 1.e-09, 1.e-09, 1.e-09, 1.e-09, 1.e-09, 1.e-09, 1.e-09, 1.e-09, 1.e-09, 1.e-09, 1.e-09]) - salt_to_mmday(time)float643.154e+05 3.154e+05 ... 3.154e+05
- long_name :
- Convert Salt to Water (millimeters/day)
array([315360., 315360., 315360., 315360., 315360., 315360., 315360., 315360., 315360., 315360., 315360., 315360.]) - momentum_factor(time)float6410.0 10.0 10.0 ... 10.0 10.0 10.0
- long_name :
- Convert Windstress to Velocity Flux
array([10., 10., 10., 10., 10., 10., 10., 10., 10., 10., 10., 10.])
- hflux_factor(time)float642.439e-05 2.439e-05 ... 2.439e-05
- long_name :
- Convert Heat and Solar Flux to Temperature Flux
array([2.43908626e-05, 2.43908626e-05, 2.43908626e-05, 2.43908626e-05, 2.43908626e-05, 2.43908626e-05, 2.43908626e-05, 2.43908626e-05, 2.43908626e-05, 2.43908626e-05, 2.43908626e-05, 2.43908626e-05]) - fwflux_factor(time)float640.0001 0.0001 ... 0.0001 0.0001
- long_name :
- Convert Net Fresh Water Flux to Salt Flux (in model units)
array([0.0001, 0.0001, 0.0001, 0.0001, 0.0001, 0.0001, 0.0001, 0.0001, 0.0001, 0.0001, 0.0001, 0.0001]) - salinity_factor(time)float64-0.00347 -0.00347 ... -0.00347
array([-0.00347, -0.00347, -0.00347, -0.00347, -0.00347, -0.00347, -0.00347, -0.00347, -0.00347, -0.00347, -0.00347, -0.00347]) - sflux_factor(time)float640.1 0.1 0.1 0.1 ... 0.1 0.1 0.1 0.1
- long_name :
- Convert Salt Flux to Salt Flux (in model units)
array([0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1, 0.1])
- nsurface_t(time)float648.61e+04 8.61e+04 ... 8.61e+04
- long_name :
- Number of Ocean T Points at Surface
array([86096., 86096., 86096., 86096., 86096., 86096., 86096., 86096., 86096., 86096., 86096., 86096.]) - nsurface_u(time)float648.297e+04 8.297e+04 ... 8.297e+04
- long_name :
- Number of Ocean U Points at Surface
array([82966., 82966., 82966., 82966., 82966., 82966., 82966., 82966., 82966., 82966., 82966., 82966.]) - UVEL(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Velocity in grid-x direction
- units :
- centimeter/s
- grid_loc :
- 3221
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - UVEL2(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Velocity**2 in grid-x direction
- units :
- centimeter^2/s^2
- grid_loc :
- 3221
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VVEL(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Velocity in grid-y direction
- units :
- centimeter/s
- grid_loc :
- 3221
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VVEL2(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Velocity**2 in grid-y direction
- units :
- centimeter^2/s^2
- grid_loc :
- 3221
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TEMP(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Potential Temperature
- units :
- degC
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - dTEMP_POS_2D(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- max pos column temperature timestep diff
- units :
- degC
- grid_loc :
- 2110
- cell_methods :
- time: maximum
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - dTEMP_NEG_2D(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- min neg column temperature timestep diff
- units :
- degC
- grid_loc :
- 2110
- cell_methods :
- time: minimum
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SALT(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Salinity
- units :
- gram/kilogram
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SSS2(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Sea Surface Salinity**2
- units :
- (gram/kilogram)**2
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TEMP2(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Temperature**2
- units :
- degC^2
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - RHO(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- In-Situ Density
- units :
- gram/centimeter^3
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - RHO_VINT(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Vertical Integral of In-Situ Density
- units :
- gram/centimeter^2
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - RESID_T(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Free-Surface Residual Flux (T)
- units :
- watt/m^2
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - RESID_S(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Free-Surface Residual Flux (S)
- units :
- kg/m^2/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SU(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Vertically Integrated Velocity in grid-x direction
- units :
- centimeter^2/s
- grid_loc :
- 2221
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SV(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Vertically Integrated Velocity in grid-y direction
- units :
- centimeter^2/s
- grid_loc :
- 2221
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SSH(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Sea Surface Height
- units :
- centimeter
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SSH2(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- SSH**2
- units :
- cm^2
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SHF(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Total Surface Heat Flux, Including SW
- units :
- watt/m^2
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SHF_QSW(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Solar Short-Wave Heat Flux
- units :
- watt/m^2
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SFWF(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Virtual Salt Flux in FW Flux formulation
- units :
- kg/m^2/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SFWF_WRST(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Virtual Salt Flux due to weak restoring
- units :
- kg/m^2/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TAUX(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Windstress in grid-x direction
- units :
- dyne/centimeter^2
- grid_loc :
- 2220
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TAUX2(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Windstress**2 in grid-x direction
- units :
- dyne^2/centimeter^4
- grid_loc :
- 2220
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TAUY(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Windstress in grid-y direction
- units :
- dyne/centimeter^2
- grid_loc :
- 2220
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TAUY2(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Windstress**2 in grid-y direction
- units :
- dyne^2/centimeter^4
- grid_loc :
- 2220
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - FW(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Freshwater Flux
- units :
- centimeter/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TFW_T(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- T flux due to freshwater flux
- units :
- watt/m^2
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TFW_S(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- S flux due to freshwater flux (kg of salt/m^2/s)
- units :
- kg/m^2/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - EVAP_F(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Evaporation Flux from Coupler
- units :
- kg/m^2/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PREC_F(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Precipitation Flux from Cpl (rain+snow)
- units :
- kg/m^2/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SNOW_F(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Snow Flux from Coupler
- units :
- kg/m^2/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - MELT_F(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Melt Flux from Coupler
- units :
- kg/m^2/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ROFF_F(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Runoff Flux from Coupler
- units :
- kg/m^2/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - IOFF_F(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Ice Runoff Flux from Coupler due to Land-Model Snow Capping
- units :
- kg/m^2/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SALT_F(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Salt Flux from Coupler (kg of salt/m^2/s)
- units :
- kg/m^2/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SENH_F(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Sensible Heat Flux from Coupler
- units :
- watt/m^2
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - LWUP_F(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Longwave Heat Flux (up) from Coupler
- units :
- watt/m^2
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - LWDN_F(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Longwave Heat Flux (dn) from Coupler
- units :
- watt/m^2
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - MELTH_F(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Melt Heat Flux from Coupler
- units :
- watt/m^2
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - IFRAC(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Ice Fraction from Coupler
- units :
- fraction
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - IAGE(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Ideal Age
- units :
- years
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TEND_TEMP(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Tendency of Thickness Weighted TEMP
- units :
- degC/s
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - RF_TEND_TEMP(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Robert Filter Tendency for TEMP
- units :
- degC/s
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TEND_SALT(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Tendency of Thickness Weighted SALT
- units :
- gram/kilogram/s
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - RF_TEND_SALT(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Robert Filter Tendency for SALT
- units :
- gram/kilogram/s
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TIDAL_DIFF(time, z_w_bot, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Jayne Tidal Diffusion
- grid_loc :
- 3113
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - QSW_HBL(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Solar Short-Wave Heat Flux in bndry layer
- units :
- watt/m^2
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - KVMIX(time, z_w_bot, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Vertical diabatic diffusivity due to Tidal Mixing + background
- units :
- centimeter^2/s
- grid_loc :
- 3113
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - KVMIX_M(time, z_w_bot, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Vertical viscosity due to Tidal Mixing + background
- units :
- centimeter^2/s
- grid_loc :
- 3113
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TPOWER(time, z_w_bot, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Energy Used by Vertical Mixing
- units :
- erg/s/cm^3
- grid_loc :
- 3113
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VDC_T(time, z_w_bot, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- total diabatic vertical TEMP diffusivity
- units :
- cm^2/s
- grid_loc :
- 3113
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VDC_S(time, z_w_bot, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- total diabatic vertical SALT diffusivity
- units :
- cm^2/s
- grid_loc :
- 3113
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VVC(time, z_w_bot, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- total vertical momentum viscosity
- units :
- cm^2/s
- grid_loc :
- 3113
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - DIA_IMPVF_TEMP(time, z_w_bot, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- TEMP Flux Across Bottom Face from Diabatic Implicit Vertical Mixing
- units :
- degC cm/s
- grid_loc :
- 3113
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - DIA_IMPVF_SALT(time, z_w_bot, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- SALT Flux Across Bottom Face from Diabatic Implicit Vertical Mixing
- units :
- gram/kilogram cm/s
- grid_loc :
- 3113
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - KAPPA_ISOP(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Isopycnal diffusion coefficient
- units :
- cm^2/s
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - KAPPA_THIC(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Thickness diffusion coefficient
- units :
- cm^2/s
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - HOR_DIFF(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Horizontal diffusion coefficient
- units :
- cm^2/s
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - DIA_DEPTH(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Depth of the Diabatic Region at the Surface
- units :
- cm
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TLT(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Transition Layer Thickness
- units :
- cm
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - INT_DEPTH(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Depth at which the Interior Region Starts
- units :
- cm
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - UISOP(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Bolus Velocity in grid-x direction (diagnostic)
- units :
- cm/s
- grid_loc :
- 3211
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VISOP(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Bolus Velocity in grid-y direction (diagnostic)
- units :
- cm/s
- grid_loc :
- 3121
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - WISOP(time, z_w_top, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Vertical Bolus Velocity (diagnostic)
- units :
- cm/s
- grid_loc :
- 3112
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ADVT_ISOP(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Vertically-Integrated T Eddy-Induced Advection Tendency (diagnostic)
- units :
- cm degC/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ADVS_ISOP(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Vertically-Integrated S Eddy-Induced Advection Tendency (diagnostic)
- units :
- cm gram/kilogram/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VNT_ISOP(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Heat Flux Tendency in grid-y Dir due to Eddy-Induced Vel (diagnostic)
- units :
- degC/s
- grid_loc :
- 3121
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VNS_ISOP(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Salt Flux Tendency in grid-y Dir due to Eddy-Induced Vel (diagnostic)
- units :
- gram/kilogram/s
- grid_loc :
- 3121
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ISOP_ADV_TEND_TEMP(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Eddy-induced advective tendency for TEMP
- units :
- degC/s
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - Redi_TEND_TEMP(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Redi tendency for TEMP
- units :
- degC/s
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ISOP_ADV_TEND_SALT(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Eddy-induced advective tendency for SALT
- units :
- gram/kilogram/s
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - Redi_TEND_SALT(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Redi tendency for SALT
- units :
- gram/kilogram/s
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - USUBM(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Submeso velocity in grid-x direction (diagnostic)
- units :
- cm/s
- grid_loc :
- 3211
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VSUBM(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Submeso velocity in grid-y direction (diagnostic)
- units :
- cm/s
- grid_loc :
- 3121
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - WSUBM(time, z_w_top, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Vertical submeso velocity (diagnostic)
- units :
- cm/s
- grid_loc :
- 3112
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - HLS_SUBM(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Horizontal length scale used in submeso
- units :
- cm
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ADVT_SUBM(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Vertically-Integrated T submeso Advection Tendency (diagnostic)
- units :
- cm degC/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ADVS_SUBM(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Vertically-Integrated S submeso Advection Tendency (diagnostic)
- units :
- cm gram/kilogram/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VNT_SUBM(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Heat Flux Tendency in grid-y Dir due to submeso Vel (diagnostic)
- units :
- degC/s
- grid_loc :
- 3121
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VNS_SUBM(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Salt Flux Tendency in grid-y Dir due to submeso Vel (diagnostic)
- units :
- gram/kilogram/s
- grid_loc :
- 3121
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SUBM_ADV_TEND_TEMP(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Submeso advective tendency for TEMP
- units :
- degC/s
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - SUBM_ADV_TEND_SALT(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Submeso advective tendency for SALT
- units :
- gram/kilogram/s
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - HDIFT(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Vertically Integrated Horz Mix T tendency
- units :
- centimeter degC/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - HDIFS(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Vertically Integrated Horz Diff S tendency
- units :
- centimeter gram/kilogram/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - WVEL(time, z_w_top, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Vertical Velocity
- units :
- centimeter/s
- grid_loc :
- 3112
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - WVEL2(time, z_w_top, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Vertical Velocity**2
- units :
- centimeter^2/s^2
- grid_loc :
- 3112
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - UET(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Flux of Heat in grid-x direction
- units :
- degC/s
- grid_loc :
- 3211
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VNT(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Flux of Heat in grid-y direction
- units :
- degC/s
- grid_loc :
- 3121
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - WTT(time, z_w_top, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Heat Flux Across Top Face
- units :
- degC/s
- grid_loc :
- 3112
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - UES(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Salt Flux in grid-x direction
- units :
- gram/kilogram/s
- grid_loc :
- 3211
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - VNS(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Salt Flux in grid-y direction
- units :
- gram/kilogram/s
- grid_loc :
- 3121
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - WTS(time, z_w_top, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Salt Flux Across Top Face
- units :
- gram/kilogram/s
- grid_loc :
- 3112
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ADVT(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Vertically-Integrated T Advection Tendency
- units :
- centimeter degC/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ADVS(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Vertically-Integrated S Advection Tendency
- units :
- centimeter gram/kilogram/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ADV_3D_TEMP(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- TEMP Advection Tendency
- units :
- degC/s
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - ADV_3D_SALT(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- SALT Advection Tendency
- units :
- gram/kilogram/s
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PV(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Potential Vorticity
- units :
- 1/s/cm
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - Q(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Static Stability (d(rho(p_r))/dz)
- units :
- gram/centimeter^4
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - PD(time, z_t, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Potential Density Ref to Surface
- units :
- gram/centimeter^3
- grid_loc :
- 3111
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - QSW_HTP(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Solar Short-Wave Heat Flux in top layer
- units :
- watt/m^2
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - QSW_3D(time, z_w_top, nlat, nlon)float32dask.array<chunksize=(1, 60, 384, 320), meta=np.ndarray>
- long_name :
- Solar Short-Wave Heat Flux
- units :
- watt/m^2
- grid_loc :
- 3112
- cell_methods :
- time: mean
Array Chunk Bytes 337.50 MiB 28.12 MiB Shape (12, 60, 384, 320) (1, 60, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - S_FLUX_ROFF_VSF_SRF(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Surface Salt Virtual Salt Flux Associated with Rivers
- units :
- g/kg*cm/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - T_FLUX_EXCH_INTRF(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Vertical Temperature Flux Across Upper/Lower Layer Interface (From EBM)
- units :
- degC*cm/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - S_FLUX_EXCH_INTRF(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Vertical Salt Flux Across Upper/Lower Layer Interface (FromEBM)
- units :
- g/kg*cm/s
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - QFLUX(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Internal Ocean Heat Flux Due to Ice Formation; heat of fusion > 0 or ice-melting potential < 0
- units :
- Watts/meter^2
- grid_loc :
- 2111
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - HMXL(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Mixed-Layer Depth
- units :
- centimeter
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - HMXL_DR(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Mixed-Layer Depth (density)
- units :
- centimeter
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - HMXL_DR2(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Mixed-Layer Depth squared (density)
- units :
- centimeter^2
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - XMXL(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Maximum Mixed-Layer Depth
- units :
- centimeter
- grid_loc :
- 2110
- cell_methods :
- time: maximum
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - XMXL_DR(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Maximum Mixed-Layer Depth (density)
- units :
- centimeter
- grid_loc :
- 2110
- cell_methods :
- time: maximum
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TMXL(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Minimum Mixed-Layer Depth
- units :
- centimeter
- grid_loc :
- 2110
- cell_methods :
- time: minimum
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TMXL_DR(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Minimum Mixed-Layer Depth (density)
- units :
- centimeter
- grid_loc :
- 2110
- cell_methods :
- time: minimum
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - HBLT(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Boundary-Layer Depth
- units :
- centimeter
- grid_loc :
- 2110
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - XBLT(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Maximum Boundary-Layer Depth
- units :
- centimeter
- grid_loc :
- 2110
- cell_methods :
- time: maximum
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - TBLT(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Minimum Boundary-Layer Depth
- units :
- centimeter
- grid_loc :
- 2110
- cell_methods :
- time: minimum
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - BSF(time, nlat, nlon)float32dask.array<chunksize=(1, 384, 320), meta=np.ndarray>
- long_name :
- Diagnostic barotropic streamfunction
- units :
- Sv
- grid_loc :
- 2220
- cell_methods :
- time: mean
Array Chunk Bytes 5.62 MiB 480.00 kiB Shape (12, 384, 320) (1, 384, 320) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - MOC(time, transport_reg, moc_comp, moc_z, lat_aux_grid)float32dask.array<chunksize=(1, 2, 3, 61, 395), meta=np.ndarray>
- long_name :
- Meridional Overturning Circulation
- units :
- Sverdrups
- cell_methods :
- time: mean
Array Chunk Bytes 6.62 MiB 564.73 kiB Shape (12, 2, 3, 61, 395) (1, 2, 3, 61, 395) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - N_HEAT(time, transport_reg, transport_comp, lat_aux_grid)float32dask.array<chunksize=(1, 2, 5, 395), meta=np.ndarray>
- long_name :
- Northward Heat Transport
- units :
- Pwatt
- cell_methods :
- time: mean
Array Chunk Bytes 185.16 kiB 15.43 kiB Shape (12, 2, 5, 395) (1, 2, 5, 395) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray - N_SALT(time, transport_reg, transport_comp, lat_aux_grid)float32dask.array<chunksize=(1, 2, 5, 395), meta=np.ndarray>
- long_name :
- Northward Salt Transport
- units :
- gram centimeter^3/kg/s
- cell_methods :
- time: mean
Array Chunk Bytes 185.16 kiB 15.43 kiB Shape (12, 2, 5, 395) (1, 2, 5, 395) Dask graph 12 chunks in 25 graph layers Data type float32 numpy.ndarray
- timePandasIndex
PandasIndex(CFTimeIndex([0001-01-17 00:00:00, 0001-02-14 00:00:00, 0001-03-17 00:00:00, 0001-04-16 00:00:00, 0001-05-17 00:00:00, 0001-06-16 00:00:00, 0001-07-17 00:00:00, 0001-08-17 00:00:00, 0001-09-16 00:00:00, 0001-10-17 00:00:00, 0001-11-16 00:00:00, 0001-12-17 00:00:00], dtype='object', length=12, calendar='noleap', freq=None)) - z_tPandasIndex
PandasIndex(Index([ 500.0, 1500.0, 2500.0, 3500.0, 4500.0, 5500.0, 6500.0, 7500.0, 8500.0, 9500.0, 10500.0, 11500.0, 12500.0, 13500.0, 14500.0, 15500.0, 16509.83984375, 17547.904296875, 18629.126953125, 19766.02734375, 20971.138671875, 22257.828125, 23640.8828125, 25137.015625, 26765.419921875, 28548.365234375, 30511.921875, 32686.798828125, 35109.34765625, 37822.76171875, 40878.46484375, 44337.76953125, 48273.671875, 52772.80078125, 57937.2890625, 63886.26171875, 70756.328125, 78700.25, 87882.5234375, 98470.5859375, 110620.421875, 124456.6875, 140049.71875, 157394.640625, 176400.328125, 196894.421875, 218645.65625, 241397.15625, 264900.125, 288938.46875, 313340.46875, 337979.34375, 362767.03125, 387645.1875, 412576.8125, 437539.25, 462519.03125, 487508.34375, 512502.8125, 537500.0], dtype='float32', name='z_t')) - z_t_150mPandasIndex
PandasIndex(Index([ 500.0, 1500.0, 2500.0, 3500.0, 4500.0, 5500.0, 6500.0, 7500.0, 8500.0, 9500.0, 10500.0, 11500.0, 12500.0, 13500.0, 14500.0], dtype='float32', name='z_t_150m')) - z_wPandasIndex
PandasIndex(Index([ 0.0, 1000.0, 2000.0, 3000.0, 4000.0, 5000.0, 6000.0, 7000.0, 8000.0, 9000.0, 10000.0, 11000.0, 12000.0, 13000.0, 14000.0, 15000.0, 16000.0, 17019.681640625, 18076.12890625, 19182.125, 20349.931640625, 21592.34375, 22923.3125, 24358.453125, 25915.580078125, 27615.259765625, 29481.470703125, 31542.373046875, 33831.2265625, 36387.47265625, 39258.046875, 42498.88671875, 46176.65625, 50370.6875, 55174.91015625, 60699.66796875, 67072.859375, 74439.8046875, 82960.6953125, 92804.3515625, 104136.8203125, 117104.015625, 131809.359375, 148290.078125, 166499.203125, 186301.4375, 207487.390625, 229803.90625, 252990.40625, 276809.84375, 301067.0625, 325613.84375, 350344.875, 375189.1875, 400101.15625, 425052.46875, 450026.0625, 475012.0, 500004.6875, 525000.9375], dtype='float32', name='z_w')) - z_w_topPandasIndex
PandasIndex(Index([ 0.0, 1000.0, 2000.0, 3000.0, 4000.0, 5000.0, 6000.0, 7000.0, 8000.0, 9000.0, 10000.0, 11000.0, 12000.0, 13000.0, 14000.0, 15000.0, 16000.0, 17019.681640625, 18076.12890625, 19182.125, 20349.931640625, 21592.34375, 22923.3125, 24358.453125, 25915.580078125, 27615.259765625, 29481.470703125, 31542.373046875, 33831.2265625, 36387.47265625, 39258.046875, 42498.88671875, 46176.65625, 50370.6875, 55174.91015625, 60699.66796875, 67072.859375, 74439.8046875, 82960.6953125, 92804.3515625, 104136.8203125, 117104.015625, 131809.359375, 148290.078125, 166499.203125, 186301.4375, 207487.390625, 229803.90625, 252990.40625, 276809.84375, 301067.0625, 325613.84375, 350344.875, 375189.1875, 400101.15625, 425052.46875, 450026.0625, 475012.0, 500004.6875, 525000.9375], dtype='float32', name='z_w_top')) - z_w_botPandasIndex
PandasIndex(Index([ 1000.0, 2000.0, 3000.0, 4000.0, 5000.0, 6000.0, 7000.0, 8000.0, 9000.0, 10000.0, 11000.0, 12000.0, 13000.0, 14000.0, 15000.0, 16000.0, 17019.681640625, 18076.12890625, 19182.125, 20349.931640625, 21592.34375, 22923.3125, 24358.453125, 25915.580078125, 27615.259765625, 29481.470703125, 31542.373046875, 33831.2265625, 36387.47265625, 39258.046875, 42498.88671875, 46176.65625, 50370.6875, 55174.91015625, 60699.66796875, 67072.859375, 74439.8046875, 82960.6953125, 92804.3515625, 104136.8203125, 117104.015625, 131809.359375, 148290.078125, 166499.203125, 186301.4375, 207487.390625, 229803.90625, 252990.40625, 276809.84375, 301067.0625, 325613.84375, 350344.875, 375189.1875, 400101.15625, 425052.46875, 450026.0625, 475012.0, 500004.6875, 525000.9375, 549999.0625], dtype='float32', name='z_w_bot')) - lat_aux_gridPandasIndex
PandasIndex(Index([-79.48815155029297, -78.95289611816406, -78.41865539550781, -77.88441467285156, -77.35017395019531, -76.81593322753906, -76.28169250488281, -75.74745178222656, -75.21321868896484, -74.6789779663086, ... 85.26966857910156, 85.79525756835938, 86.32085418701172, 86.84644317626953, 87.37203979492188, 87.89762878417969, 88.42322540283203, 88.94881439208984, 89.47441101074219, 90.0], dtype='float32', name='lat_aux_grid', length=395)) - moc_zPandasIndex
PandasIndex(Index([ 0.0, 1000.0, 2000.0, 3000.0, 4000.0, 5000.0, 6000.0, 7000.0, 8000.0, 9000.0, 10000.0, 11000.0, 12000.0, 13000.0, 14000.0, 15000.0, 16000.0, 17019.681640625, 18076.12890625, 19182.125, 20349.931640625, 21592.34375, 22923.3125, 24358.453125, 25915.580078125, 27615.259765625, 29481.470703125, 31542.373046875, 33831.2265625, 36387.47265625, 39258.046875, 42498.88671875, 46176.65625, 50370.6875, 55174.91015625, 60699.66796875, 67072.859375, 74439.8046875, 82960.6953125, 92804.3515625, 104136.8203125, 117104.015625, 131809.359375, 148290.078125, 166499.203125, 186301.4375, 207487.390625, 229803.90625, 252990.40625, 276809.84375, 301067.0625, 325613.84375, 350344.875, 375189.1875, 400101.15625, 425052.46875, 450026.0625, 475012.0, 500004.6875, 525000.9375, 549999.0625], dtype='float32', name='moc_z'))
- title :
- b.e21.B1850.f19_g17.piControl.001
- history :
- none
- Conventions :
- CF-1.0; http://www.cgd.ucar.edu/cms/eaton/netcdf/CF-current.htm
- time_period_freq :
- month_1
- model_doi_url :
- https://doi.org/10.5065/D67H1H0V
- contents :
- Diagnostic and Prognostic Variables
- source :
- CCSM POP2, the CCSM Ocean Component
- revision :
- $Id$
- calendar :
- All years have exactly 365 days.
- start_time :
- This dataset was created on 2025-06-14 at 12:35:19.2
- cell_methods :
- cell_methods = time: mean ==> the variable values are averaged over the time interval between the previous time coordinate and the current one. cell_methods absent ==> the variable values are at the time given by the current time coordinate.
Calculate annual mean SST and make a simple plot using Xarray#
sst_PI = ds_PI_ocn.TEMP.isel(z_t=0).mean('time')
sst_MH = ds_MH_ocn.TEMP.isel(z_t=0).mean('time')
sst_PI.plot(size=1.5)
<matplotlib.collections.QuadMesh at 0x14895d798590>
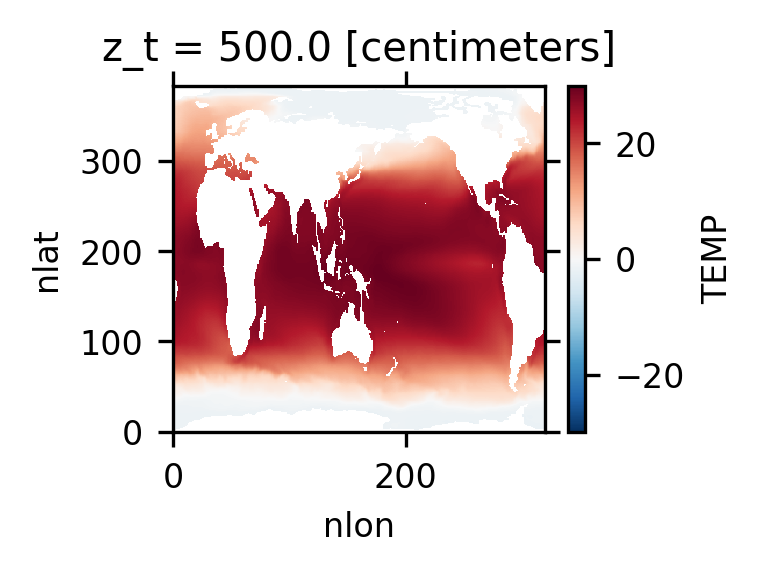
What do nlat and nlon mean?
Where is Greenland? It is the “north pole” in the model!
Figure: POP ocean grid
Regridding is needed!#
Use the
RegridderfromxesmfUse the
util.grid_globalto create a 1x1 regular gridUse linear interpolation
Use IPython magic command to
timethe runtime
%%time
ds_PI_ocn['lat'] = ds_PI_ocn.TLAT
ds_PI_ocn['lon'] = ds_PI_ocn.TLONG
regridder = xesmf.Regridder(
ds_in=ds_PI_ocn,
ds_out=xesmf.util.grid_global(1, 1, cf=True, lon1=360),
method='bilinear',
periodic=True)
sst_PI_1x1 = regridder(sst_PI)
sst_MH_1x1 = regridder(sst_MH)
sst_PI_1x1
CPU times: user 4.79 s, sys: 279 ms, total: 5.07 s
Wall time: 5.64 s
<xarray.DataArray (lat: 180, lon: 360)> Size: 259kB
dask.array<astype, shape=(180, 360), dtype=float32, chunksize=(180, 360), chunktype=numpy.ndarray>
Coordinates:
z_t float32 4B 500.0
* lon (lon) float64 3kB 0.5 1.5 2.5 3.5 ... 357.5 358.5 359.5
latitude_longitude float64 8B nan
* lat (lat) float64 1kB -89.5 -88.5 -87.5 ... 87.5 88.5 89.5
Attributes:
regrid_method: bilinear- lat: 180
- lon: 360
- dask.array<chunksize=(180, 360), meta=np.ndarray>
Array Chunk Bytes 253.12 kiB 253.12 kiB Shape (180, 360) (180, 360) Dask graph 1 chunks in 35 graph layers Data type float32 numpy.ndarray - z_t()float32500.0
array(500., dtype=float32)
- lon(lon)float640.5 1.5 2.5 ... 357.5 358.5 359.5
- bounds :
- lon_bounds
- standard_name :
- longitude
- units :
- degrees_east
array([ 0.5, 1.5, 2.5, ..., 357.5, 358.5, 359.5])
- latitude_longitude()float64nan
array(nan)
- lat(lat)float64-89.5 -88.5 -87.5 ... 88.5 89.5
- bounds :
- lat_bounds
- standard_name :
- latitude
- units :
- degrees_north
array([-89.5, -88.5, -87.5, -86.5, -85.5, -84.5, -83.5, -82.5, -81.5, -80.5, -79.5, -78.5, -77.5, -76.5, -75.5, -74.5, -73.5, -72.5, -71.5, -70.5, -69.5, -68.5, -67.5, -66.5, -65.5, -64.5, -63.5, -62.5, -61.5, -60.5, -59.5, -58.5, -57.5, -56.5, -55.5, -54.5, -53.5, -52.5, -51.5, -50.5, -49.5, -48.5, -47.5, -46.5, -45.5, -44.5, -43.5, -42.5, -41.5, -40.5, -39.5, -38.5, -37.5, -36.5, -35.5, -34.5, -33.5, -32.5, -31.5, -30.5, -29.5, -28.5, -27.5, -26.5, -25.5, -24.5, -23.5, -22.5, -21.5, -20.5, -19.5, -18.5, -17.5, -16.5, -15.5, -14.5, -13.5, -12.5, -11.5, -10.5, -9.5, -8.5, -7.5, -6.5, -5.5, -4.5, -3.5, -2.5, -1.5, -0.5, 0.5, 1.5, 2.5, 3.5, 4.5, 5.5, 6.5, 7.5, 8.5, 9.5, 10.5, 11.5, 12.5, 13.5, 14.5, 15.5, 16.5, 17.5, 18.5, 19.5, 20.5, 21.5, 22.5, 23.5, 24.5, 25.5, 26.5, 27.5, 28.5, 29.5, 30.5, 31.5, 32.5, 33.5, 34.5, 35.5, 36.5, 37.5, 38.5, 39.5, 40.5, 41.5, 42.5, 43.5, 44.5, 45.5, 46.5, 47.5, 48.5, 49.5, 50.5, 51.5, 52.5, 53.5, 54.5, 55.5, 56.5, 57.5, 58.5, 59.5, 60.5, 61.5, 62.5, 63.5, 64.5, 65.5, 66.5, 67.5, 68.5, 69.5, 70.5, 71.5, 72.5, 73.5, 74.5, 75.5, 76.5, 77.5, 78.5, 79.5, 80.5, 81.5, 82.5, 83.5, 84.5, 85.5, 86.5, 87.5, 88.5, 89.5])
- lonPandasIndex
PandasIndex(Index([ 0.5, 1.5, 2.5, 3.5, 4.5, 5.5, 6.5, 7.5, 8.5, 9.5, ... 350.5, 351.5, 352.5, 353.5, 354.5, 355.5, 356.5, 357.5, 358.5, 359.5], dtype='float64', name='lon', length=360)) - latPandasIndex
PandasIndex(Index([-89.5, -88.5, -87.5, -86.5, -85.5, -84.5, -83.5, -82.5, -81.5, -80.5, ... 80.5, 81.5, 82.5, 83.5, 84.5, 85.5, 86.5, 87.5, 88.5, 89.5], dtype='float64', name='lat', length=180))
- regrid_method :
- bilinear
lat = sst_PI_1x1.lat
lon = sst_PI_1x1.lon
fig, axes = plt.subplots(nrows=1, ncols=3,
figsize=(10, 2),
subplot_kw={'projection': ccrs.Robinson(central_longitude=210)},
constrained_layout=True)
ax = axes[0]
ax.set_title("PI SST, annual mean")
sst_PI_new, lon_new = add_cyclic_point(sst_PI_1x1, lon)
p0 = ax.contourf(lon_new, lat, sst_PI_new,
levels=np.linspace(-2, 30, 17),
cmap='inferno', extend='both',
transform=ccrs.PlateCarree())
plt.colorbar(p0, ax=ax)
ax = axes[1]
ax.set_title("MH SST, annual mean")
sst_MH_new, lon_new = add_cyclic_point(sst_MH_1x1, lon)
p1 = ax.contourf(lon_new, lat, sst_MH_new,
levels=np.linspace(-2, 30, 17),
cmap='inferno', extend='both',
transform=ccrs.PlateCarree())
plt.colorbar(p1, ax=ax)
ax = axes[2]
ax.set_title("MH-PI SST")
p2 = ax.contourf(lon_new, lat, sst_MH_new - sst_PI_new,
cmap='coolwarm',
levels=np.linspace(-2, 2, 21),
extend='both',
transform=ccrs.PlateCarree())
plt.colorbar(p2, ax=ax)
for ax in axes:
ax.set_global()
ax.coastlines(linewidth=0.5)
# We could use savefig to save the plot as pdf
# plt.savefig('SST_xy.PI_vs_MH.pdf', format='pdf', bbox_inches="tight")

Small group discussion#
Colder SSTs over lots of the regions? Internal variability?
We need to consider internal variability and test significance
What’s the timescale of upper ocean response?
Summary so far#
Load multiple CESM output using
XarrayPlot variables in zonal mean and map view
Interpolate the atmosphere vertical coordinate from hybrid to pressure is needed!
Ocean models usually use irregular grid with poles moved into land; regridding is needed!
Mid-Holocene orbital forcing is seasonal and impacts atmosphere circulation, precipitation, and other climate states.
